7ZXU
 
 | | SARS-CoV-2 Omicron BA.4/5 RBD in complex with Beta-27 Fab and C1 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-27 heavy chain, Beta-27 light chain, ... | | Authors: | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-23 | | Release date: | 2022-06-29 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum.
Cell, 185, 2022
|
|
6GX3
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with an hydroxamate 1 | | Descriptor: | 4-chloranyl-~{N}-oxidanyl-1-benzothiophene-2-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis, Crystallization Studies, and in vitro Characterization of Cinnamic Acid Derivatives as SmHDAC8 Inhibitors for the Treatment of Schistosomiasis.
ChemMedChem, 13, 2018
|
|
5ZT4
 
 | |
7QLB
 
 | | SMYD3 in complex with fragment FL06268 | | Descriptor: | 1-methylimidazole-4-sulfonamide, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYLMETHIONINE, ... | | Authors: | Lund, B.A, Cederfelt, D, Dobritzsch, D. | | Deposit date: | 2021-12-20 | | Release date: | 2023-03-29 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of fragments targeting SMYD3 using highly sensitive kinetic and multiplexed biosensor-based screening
Rsc Med Chem, 2024
|
|
8A9I
 
 | | PI3KC2a core in complex with PITCOIN1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-oxidanylidene-3-(2-phenylethyl)pteridin-2-yl]sulfanyl-~{N}-(1,3-thiazol-2-yl)ethanamide, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, ... | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2022-06-28 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Development of selective inhibitors of phosphatidylinositol 3-kinase C2 alpha.
Nat.Chem.Biol., 19, 2023
|
|
6GNH
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and an Azepanyl Phenyl Benzylsulphonamide Ligand | | Descriptor: | Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, methyl 4-(azepan-1-yl)-3-[(4-methoxyphenyl)sulfonylamino]benzoate | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
9FAI
 
 | |
6GVA
 
 | | CDK2/cyclin A2 in complex with pyrazolo[4,3-d]pyrimidine inhibitor LGR4455 | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-azanylethylsulfanyl)-3-propan-2-yl-~{N}-[(4-pyridin-2-ylphenyl)methyl]-2~{H}-pyrazolo[4,3-d]pyrimidin-7-amine, BROMIDE ION, ... | | Authors: | Skerlova, J, Rezacova, P. | | Deposit date: | 2018-06-20 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3,5,7-Substituted Pyrazolo[4,3- d]pyrimidine Inhibitors of Cyclin-Dependent Kinases and Their Evaluation in Lymphoma Models.
J.Med.Chem., 62, 2019
|
|
3ABN
 
 | | Crystal structure of (Pro-Pro-Gly)4-Hyp-Asp-Gly-(Pro-Pro-Gly)4 at 1.02 A | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Shimura, M, Kawaguchi, T, Noguchi, K, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2009-12-16 | | Release date: | 2010-12-01 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the collagen model peptide (Pro-Pro-Gly)4 -Hyp-Asp-Gly-(Pro-Pro-Gly)4 at 1.0 angstrom resolution.
Biopolymers, 99, 2013
|
|
8VKA
 
 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 9c | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fenwick, M.K, Staker, B.L, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Exploring Subsite Selectivity within Plasmodium vivax N -Myristoyltransferase Using Pyrazole-Derived Inhibitors.
J.Med.Chem., 67, 2024
|
|
8VKB
 
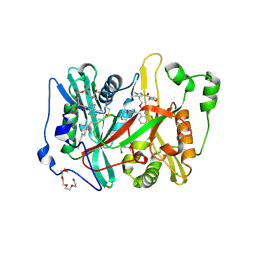 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 10b | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, ... | | Authors: | Fenwick, M.K, Staker, B.L, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Exploring Subsite Selectivity within Plasmodium vivax N -Myristoyltransferase Using Pyrazole-Derived Inhibitors.
J.Med.Chem., 67, 2024
|
|
5BSZ
 
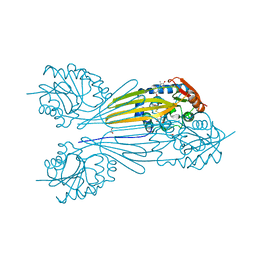 | | X-ray structure of the sugar N-methyltransferase KedS8 from Streptoalloteichus sp ATCC 53650 | | Descriptor: | 1,2-ETHANEDIOL, N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Thoden, J.B, Delvaux, N.A, Holden, H.M. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular architecture of KedS8, a sugar N-methyltransferase from Streptoalloteichus sp. ATCC 53650.
Protein Sci., 24, 2015
|
|
8VVM
 
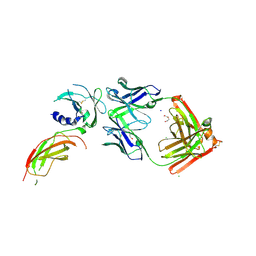 | | Structure of FabS1CE1-EPR1-1 in complex with the erythropoietin receptor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
1EN1
 
 | | STRUCTURE OF THE HIV-1 MINUS STRAND PRIMER BINDING SITE | | Descriptor: | DNA (5'-D(P*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3') | | Authors: | Johnson, P.E, Turner, R.B, Wu, Z.R, Levin, J.G, Summers, M.F. | | Deposit date: | 2000-03-20 | | Release date: | 2000-04-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription
Biochemistry, 39, 2000
|
|
5L6K
 
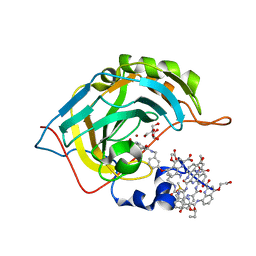 | | Crystal Structure of Human Carbonic Anhydrase II in Complex with a Quinoline Oligoamide Foldamer | | Descriptor: | Aromatic foldamer, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Jewginski, M, Langlois d'Estaintot, B, Granier, T, Huc, Y. | | Deposit date: | 2016-05-30 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Self-Assembled Protein-Aromatic Foldamer Complexes with 2:3 and 2:2:1 Stoichiometries.
J. Am. Chem. Soc., 139, 2017
|
|
7B0W
 
 | | Crystal structure of the E. coli type 1 pilus assembly inhibitor FimI bound to FimC | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein FimC, FORMIC ACID, ... | | Authors: | Scharer, M.A, Zigova, Z, Giese, C, Puorger, C, Ignatov, O, Capitani, G, Glockshuber, R. | | Deposit date: | 2020-11-23 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
2Q9D
 
 | | Structure of spin-labeled T4 lysozyme mutant A41R1 | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-06-12 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | EPR (1.4 Å), X-RAY DIFFRACTION | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme.
Protein Sci., 17, 2008
|
|
5C5G
 
 | |
5ZVR
 
 | |
4YEP
 
 | | L4b Domain of Human Laminin alpha-2 | | Descriptor: | 1,2-ETHANEDIOL, Laminin subunit alpha-2 | | Authors: | Toot, M, Gat, Y, Fass, D. | | Deposit date: | 2015-02-24 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Laminin L4 domain structure resembles adhesion modules in ephrin receptor and other transmembrane glycoproteins.
Febs J., 282, 2015
|
|
7E4F
 
 | | Mycobacterium tuberculosis enolase mutant - E204A complex with phosphoenolpyruvate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ahmad, M, Pal, R.K, Biswal, B.K. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural snapshots of Mycobacterium tuberculosis enolase reveal dual mode of 2PG binding and its implication in enzyme catalysis.
Iucrj, 10, 2023
|
|
4LY0
 
 | | Crystal structure WlaRD, a sugar 3N-formyl transferase in the presence of dTDP-Glc and 10-N-Formyl-THF | | Descriptor: | 1,2-ETHANEDIOL, 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
8VTP
 
 | | Structure of FabS1CE-EPR-1, a high affinity antibody for the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, AMMONIUM ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
7UJ2
 
 | | OspC Type B | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Outer surface protein C | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2022-03-30 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural Elucidation of a Protective B Cell Epitope on Outer Surface Protein C (OspC) of the Lyme Disease Spirochete, Borreliella burgdorferi.
Mbio, 14, 2023
|
|
6A1C
 
 | | Crystal structure of the CK2a1-go289 complex | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-4-[(E)-(3-methylsulfanyl-5-phenyl-1,2,4-triazol-4-yl)iminomethyl]phenol, Casein kinase II subunit alpha, ... | | Authors: | Kinoshita, T, Tsuyuguchi, M. | | Deposit date: | 2018-06-07 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth.
Sci Adv, 5, 2019
|
|
