1X92
 
 | | CRYSTAL STRUCTURE OF PSEUDOMONAS AERUGINOSA PHOSPHOHEPTOSE ISOMERASE IN COMPLEX WITH REACTION PRODUCT D-GLYCERO-D-MANNOPYRANOSE-7-PHOSPHATE | | Descriptor: | 7-O-phosphono-D-glycero-alpha-D-manno-heptopyranose, PHOSPHOHEPTOSE ISOMERASE | | Authors: | Walker, J.R, Evdokimova, E, Kudritska, M, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-19 | | Release date: | 2004-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of sedoheptulose-7-phosphate isomerase, a critical enzyme for lipopolysaccharide biosynthesis and a target for antibiotic adjuvants.
J.Biol.Chem., 283, 2008
|
|
2VRC
 
 | | Crystal structure of the Citrobacter sp. triphenylmethane reductase complexed with NADP(H) | | Descriptor: | TRIPHENYLMETHANE REDUCTASE | | Authors: | Kim, Y, Park, H.J, Kwak, S.N, Lee, J.S, Oh, T.K, Kim, M.H. | | Deposit date: | 2008-03-31 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insight Into Bioremediation of Triphenylmethane Dyes by Citrobacter Sp. Triphenylmethane Reductase.
J.Biol.Chem., 283, 2008
|
|
6HTG
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 4 | | Descriptor: | 3-benzamido-4-chloranyl-~{N}-oxidanyl-benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.939 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HTZ
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 8 | | Descriptor: | 4-methoxy-~{N}-oxidanyl-3-(2-phenylethanoylamino)benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
8E4X
 
 | |
8E0F
 
 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2-RD) bound to dsRNA containing a G-G pair adjacent to the target site | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5-R(*GP*CP*UP*CP*GP*CP*GP*AP*UP*GP*CP*GP*(8AZ)P*GP*AP*GP*GP*GP*CP* UP*CP*UP*GP*AP*UP*AP*GP*CP*UP*AP*CP*G)-3), ... | | Authors: | Wilcox, X.E, Fisher, A.J, Beal, P.A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ADAR activation by inducing a syn conformation at guanosine adjacent to an editing site.
Nucleic Acids Res., 50, 2022
|
|
1YLA
 
 | | Ubiquitin-conjugating enzyme E2-25 kDa (Huntington interacting protein 2) | | Descriptor: | Ubiquitin-conjugating enzyme E2-25 kDa | | Authors: | Choe, J, Avvakumov, G.V, Newman, E.M, Mackenzie, F, Kozieradzki, I, Bochkarev, A, Sundstrom, M, Arrowsmith, C, Edwards, A, Dhe-paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity
J.Biol.Chem., 285, 2010
|
|
6HSG
 
 | | Crystal structure of Schistosoma mansoni HDAC8 H292M mutant complexed with NCC-149 | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, Histone deacetylase, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HU1
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 10 | | Descriptor: | 4-chloranyl-3-[(2,4-dichlorophenyl)carbonylamino]-~{N}-oxidanyl-benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
2ZTD
 
 | | MtRuvA Form III | | Descriptor: | GLYCEROL, Holliday junction ATP-dependent DNA helicase ruvA | | Authors: | Prabu, J.R, Thamotharan, S, Khanduja, J.S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-10-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and modelling studies on Mycobacterium tuberculosis RuvA Additional role of RuvB-binding domain and inter species variability
Biochim.Biophys.Acta, 1794, 2009
|
|
6HU0
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 9 | | Descriptor: | 3-[(2,4-dichlorophenyl)carbonylamino]-4-methoxy-~{N}-oxidanyl-benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HQY
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with PCI-34051 | | Descriptor: | 1-[(4-methoxyphenyl)methyl]-~{N}-oxidanyl-indole-6-carboxamide, GLYCEROL, Histone deacetylase, ... | | Authors: | Marek, M, Shaik, T.B, Romier, C. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
8DY2
 
 | | Crystal Structure of spFv GLK1 | | Descriptor: | SULFATE ION, spFv GLK1 LH | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DY4
 
 | | Crystal Structure of spFv CAT2200 HL | | Descriptor: | spFv CAT2200 HL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DY0
 
 | | Crystal Structure of spFv GLK1 HL | | Descriptor: | spFv GLK1 HL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DY3
 
 | | Crystal Structure of spFv GLK2 HL | | Descriptor: | SULFATE ION, spFv GLK2 HL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.509 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DNY
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by decatransin | | Descriptor: | Decatransin peptide inhibitor, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DNZ
 
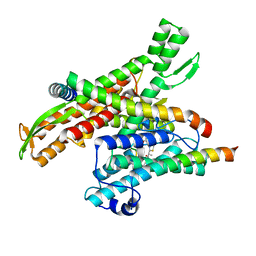 | | Cryo-EM structure of the human Sec61 complex inhibited by apratoxin F | | Descriptor: | Apratoxin F peptide inhibitor, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DO3
 
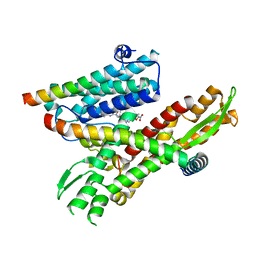 | | Cryo-EM structure of the human Sec61 complex inhibited by eeyarestatin I | | Descriptor: | N'-(4-chlorophenyl)-N-[(4R)-3-(4-chlorophenyl)-5,5-dimethyl-1-(2-{(2E)-2-[(2E)-3-(5-nitrofuran-2-yl)prop-2-en-1-ylidene]hydrazinyl}-2-oxoethyl)-2-oxoimidazolidin-4-yl]-N-hydroxyurea, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DNX
 
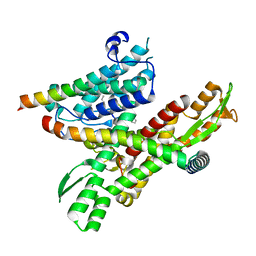 | | Cryo-EM structure of the human Sec61 complex inhibited by cotransin | | Descriptor: | Cotransin analogue peptide inhibitor, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DNV
 
 | |
8DNW
 
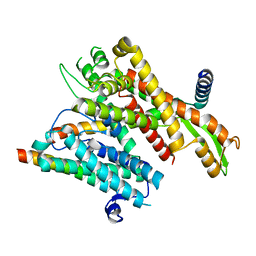 | |
8DO0
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by mycolactone | | Descriptor: | Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, Protein transport protein Sec61 subunit gamma, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DO2
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by cyclotriazadisulfonamide (CADA) | | Descriptor: | 9-benzyl-1,5-bis(4-methylbenzene-1-sulfonyl)-3-methylidene-1,5,9-triazacyclododecane, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DO1
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by ipomoeassin F | | Descriptor: | Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, Protein transport protein Sec61 subunit gamma, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
