1TPO
 
 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | BETA-TRYPSIN, CALCIUM ION | | Authors: | Bode, W, Walter, J, Huber, R. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
1TPP
 
 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | (2S)-3-(4-carbamimidoylphenyl)-2-hydroxypropanoic acid, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Walter, J, Bode, W, Huber, R. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
1TPS
 
 | |
1TPT
 
 | | THREE-DIMENSIONAL STRUCTURE OF THYMIDINE PHOSPHORYLASE FROM ESCHERICHIA COLI AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | SULFATE ION, THYMIDINE PHOSPHORYLASE, THYMINE | | Authors: | Walter, M.R, Cook, W.J, Cole, L.B, Short, S.A, Koszalka, G.W, Krenitsky, T.A, Ealick, S.E. | | Deposit date: | 1990-06-14 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of thymidine phosphorylase from Escherichia coli at 2.8 A resolution.
J.Biol.Chem., 265, 1990
|
|
1TPU
 
 | | S96P CHANGE IS A SECOND-SITE SUPPRESSOR FOR H95N SLUGGISH MUTANT TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the H95N lesion by the secondary S96P mutation in triosephosphate isomerase.
Biochemistry, 35, 1996
|
|
1TPV
 
 | | S96P CHANGE IS A SECOND-SITE SUPPRESSOR FOR H95N SLUGGISH MUTANT TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the H95N lesion by the secondary S96P mutation in triosephosphate isomerase.
Biochemistry, 35, 1996
|
|
1TPW
 
 | | TRIOSEPHOSPHATE ISOMERASE DRINKS WATER TO KEEP HEALTHY | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of water in the catalytic efficiency of triosephosphate isomerase.
Biochemistry, 38, 1999
|
|
1TPX
 
 | | Ovine recombinant PrP(114-234), ARQ variant in complex with the Fab of the VRQ14 antibody | | Descriptor: | major prion protein, the VRQ14 Fab | | Authors: | Eghiaian, F, Grosclaude, J, Lesceu, S, Debey, P, Doublet, B, Treguer, E, Rezaei, H, Knossow, M. | | Deposit date: | 2004-06-16 | | Release date: | 2004-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Insight into the PrPC -> PrPSc conversion from the structures of antibody-bound ovine prion scrapie-susceptibility variants.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TPY
 
 | |
1TPZ
 
 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1TQ0
 
 | | Crystal structure of the potent anticoagulant thrombin mutant W215A/E217A in free form | | Descriptor: | Prothrombin | | Authors: | Pineda, A.O, Chen, Z.-W, Caccia, S, Savvides, S.N, Waksman, G, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The anticoagulant thrombin mutant W215A/E217A has a collapsed primary specificity pocket
J.Biol.Chem., 279, 2004
|
|
1TQ1
 
 | | Solution structure of At5g66040, a putative protein from Arabidosis Thaliana | | Descriptor: | senescence-associated family protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Singh, S, Lee, M.S, Tyler, E.M, Shahan, M.N, Vinarov, D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-06-16 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a single-domain thiosulfate sulfurtransferase from Arabidopsis thaliana.
Protein Sci., 15, 2006
|
|
1TQ2
 
 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1TQ3
 
 | |
1TQ4
 
 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, interferon-inducible GTPase | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1TQ5
 
 | |
1TQ6
 
 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, interferon-inducible GTPase | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1TQ7
 
 | | Crystal structure of the anticoagulant thrombin mutant W215A/E217A bound to PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Prothrombin, ... | | Authors: | Pineda, A.O, Chen, Z.-W, Caccia, S, Savvides, S.N, Waksman, G, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The anticoagulant thrombin mutant W215A/E217A has a collapsed primary specificity pocket
J.Biol.Chem., 279, 2004
|
|
1TQ8
 
 | | Crystal Structure of protein Rv1636 from Mycobacterium tuberculosis H37Rv | | Descriptor: | hypothetical protein Rv1636 | | Authors: | Rajashankar, K.R, Kniewel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-16 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of hypothetical protein Rv1636 from Mycobacterium tuberculosis H37Rv
To be Published
|
|
1TQ9
 
 | | Non-covalent swapped dimer of Bovine Seminal Ribonuclease in complex with 2'-DEOXYCYTIDINE-2'-DEOXYADENOSINE-3',5'-MONOPHOSPHATE | | Descriptor: | 2'-DEOXYCYTIDINE-2'-DEOXYADENOSINE-3',5'-MONOPHOSPHATE, Ribonuclease, seminal | | Authors: | Sica, F, Di Fiore, A, Merlino, A, Mazzarella, L. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Stability of the Non-covalent Swapped Dimer of Bovine Seminal Ribonuclease: AN ENZYME TAILORED TO EVADE RIBONUCLEASE PROTEIN INHIBITOR
J.Biol.Chem., 279, 2004
|
|
1TQB
 
 | | Ovine recombinant PrP(114-234), VRQ variant in complex with the Fab of the VRQ14 antibody | | Descriptor: | VRQ14 Fab Heavy chain, VRQ14 Fab light chain, prion protein | | Authors: | Eghiaian, F, Grosclaude, J, Debey, P, Doublet, B, Treguer, E, Rezaei, H, Knossow, M. | | Deposit date: | 2004-06-17 | | Release date: | 2004-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Insight into the PrPC-->PrPSc conversion from the structures of antibody-bound ovine prion scrapie-susceptibility variants
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TQC
 
 | | Ovine recombinant PrP(114-234), ARR variant in complex with the VRQ14 Fab fragment (IgG2a) | | Descriptor: | VRQ14 Fab Heavy chain, VRQ14 Fab light chain, prion protein | | Authors: | Eghiaian, F, Grosclaude, J, Lesceu, S, Debey, P, Doublet, B, Treguer, E, Rezaei, H, Knossow, M. | | Deposit date: | 2004-06-17 | | Release date: | 2004-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insight into the PrPC-->PrPSc conversion from the structures of antibody-bound ovine prion scrapie-susceptibility variants
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TQD
 
 | | Crystal structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, interferon-inducible GTPase | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
1TQE
 
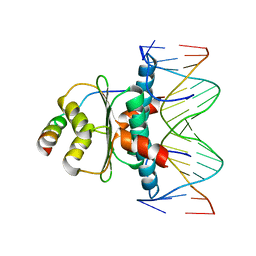 | | Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2 | | Descriptor: | Histone deacetylase 9, MEF2 binding site of nur77 promoter, Myocyte-specific enhancer factor 2B | | Authors: | Chen, L, Han, A, He, J, Wu, Y, Liu, J.O. | | Deposit date: | 2004-06-17 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Recruitment of Class II Histone Deacetylases by Myocyte Enhancer Factor-2.
J.Mol.Biol., 345, 2005
|
|
1TQF
 
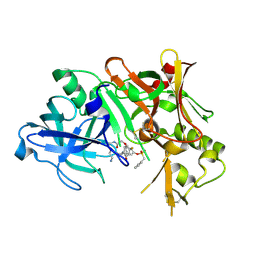 | | Crystal structure of human Beta secretase complexed with inhibitor | | Descriptor: | 3-{2-[(5-AMINOPENTYL)AMINO]-2-OXOETHOXY}-5-({[1-(4-FLUOROPHENYL)ETHYL]AMINO}CARBONYL)PHENYL PHENYLMETHANESULFONATE, Beta-secretase 1 | | Authors: | Munshi, S, Chen, Z, Kuo, L. | | Deposit date: | 2004-06-17 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a small molecule nonpeptide active site beta-secretase inhibitor that displays a nontraditional binding mode for aspartyl proteases.
J.Med.Chem., 47, 2004
|
|
