6YNX
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - Fo-subcomplex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kock Flygaard, R, Muhleip, A, Amunts, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-09-30 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization.
Nat Commun, 11, 2020
|
|
6TMO
 
 | | Structure determination of an enhanced affinity TCR, a24b17, in complex with HLA-A*02:01 presenting a MART-1 peptide, EAAGIGILTV | | Descriptor: | 1,2-ETHANEDIOL, Alpha chain of engineered high affinity T-cell receptor, Beta chain of high affinity engineered T-cell receptor, ... | | Authors: | Rizkallah, P.J, Cole, D.K. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Rules Underpinning Enhanced Affinity Binding of Human T Cell Receptors Engineered for Immunotherapy.
Mol Ther Oncolytics, 18, 2020
|
|
8G3E
 
 | | Crystal structure of human WDR5 in complex with (1M)-N-[(3,5-difluoro[1,1'-biphenyl]-4-yl)methyl]-6-methyl-4-oxo-1-(pyridin-3-yl)-1,4-dihydropyridazine-3-carboxamide (compound 2, WDR5-MYC inhibitor) | | Descriptor: | (1M)-N-[(3,5-difluoro[1,1'-biphenyl]-4-yl)methyl]-6-methyl-4-oxo-1-(pyridin-3-yl)-1,4-dihydropyridazine-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Zhao, M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery and Structure-Based Design of Inhibitors of the WD Repeat-Containing Protein 5 (WDR5)-MYC Interaction.
J.Med.Chem., 66, 2023
|
|
8RBY
 
 | |
9DQF
 
 | |
5GY8
 
 | |
5GYC
 
 | |
7RBQ
 
 | | Co-crystal structure of human PRMT9 in complex with MT556 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 7-[5-S-(4-{[(4-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Protein arginine N-methyltransferase 9, ... | | Authors: | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Co-crystal structure of human PRMT9 in complex with MT556 inhibitor
To Be Published
|
|
8RJ0
 
 | |
4RH2
 
 | |
8RLP
 
 | | Human Carbonic Anhydrase II in complex with veralipride | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 2, Veralipride, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2024-01-03 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.342 Å) | | Cite: | The dopamine D 2 receptors antagonist Veralipride inhibits carbonic anhydrases: solution and crystallographic insights on human isoforms.
Chem Asian J, 19, 2024
|
|
5GQI
 
 | | Crystal structure of Cypovirus Polyhedra mutant with deletion of Ala194 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
8D6D
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 39 | | Descriptor: | (1P)-2-amino-5-bromo-1-(3-hydroxy-2,6-dimethylphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, 1,2-ETHANEDIOL, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, ... | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
9GJQ
 
 | |
5GYA
 
 | |
5N1J
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Giastas, P, Andreou, A, Balomenou, S, Bouriotis, V, Eliopoulos, E.E. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
8FK4
 
 | | Structure of the catalytic domain of Streptococcus mutans GtfB complexed to acarbose in orthorhombic space group P21212 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, Glucosyltransferase-I, ... | | Authors: | Schormann, N, Deivanayagam, C. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The catalytic domains of Streptococcus mutans glucosyltransferases: a structural analysis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
6TUZ
 
 | | Theophylline-Notum complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Caffeine inhibits Notum activity by binding at the catalytic pocket.
Commun Biol, 3, 2020
|
|
8TV1
 
 | | Structure of the EphA2 LBDCRD bound to FabS1C_L1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ephrin type-A receptor 2, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-17 | | Release date: | 2024-06-26 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthetic antibodies targeting EphA2 induce diverse signaling-competent clusters with differential activation.
Protein Sci., 34, 2025
|
|
6Q8C
 
 | | Neisseria gonorrhoeae Leucyl-tRNA Synthetase in Complex with 5'-O-(N-(L-Leucyl)-Sulfamoyl)Uridine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-Leucyl)-Sulfamoyl)Uridine, Leucine--tRNA ligase, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Comparative analysis of pyrimidine substituted aminoacyl-sulfamoyl nucleosides as potential inhibitors targeting class I aminoacyl-tRNA synthetases.
Eur.J.Med.Chem., 173, 2019
|
|
1E4N
 
 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZMGluE191D in complex with the natural aglycone DIMBOA | | Descriptor: | 2,4-DIHYDROXY-7-(METHYLOXY)-2H-1,4-BENZOXAZIN-3(4H)-ONE, BETA-GLUCOSIDASE | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Zamboni, V, Henrissat, B, Esen, A. | | Deposit date: | 2000-07-11 | | Release date: | 2000-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Mechanism of Substrate (Aglycone) Specificity in Beta -Glucosidases is Revealed by Crystal Structures of Mutant Maize Beta -Glucosidase-Dimboa, -Dimboaglc, and -Dhurrin Complexes
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5MZ5
 
 | | Crystal structure of aldehyde dehydrogenase 21 (ALDH21) from Physcomitrella patens in its apoform | | Descriptor: | 1,2-ETHANEDIOL, ALDH21), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kopecny, D, Koncitikova, R, Briozzo, P, Morera, S. | | Deposit date: | 2017-01-30 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase.
Plant J., 92, 2017
|
|
8WXJ
 
 | | human CLC-1 D136G closed state | | Descriptor: | Chloride channel protein 1, THIOCYANATE ION | | Authors: | Zhang, M.F. | | Deposit date: | 2023-10-29 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | CLC-1 in SCN environment
To Be Published
|
|
8Z6B
 
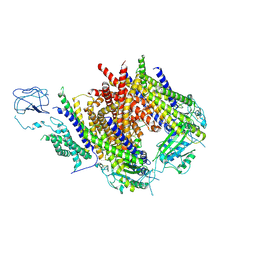 | | Structure of Polycystin-1/Polycystin-2 complex | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystin-1, ... | | Authors: | Chen, M.Y, Su, Q, Shi, Y.G. | | Deposit date: | 2024-04-18 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of polycystin1/2 complex
To Be Published
|
|
8X1G
 
 | |
