6V79
 
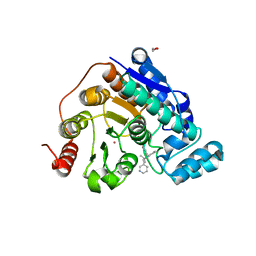 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with NF2376 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2S)-3,3-dimethyl-2-(pyridin-3-yl)-2,3-dihydro-1H-indol-1-yl]methyl}-N-hydroxybenzamide, Hdac6 protein, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03951526 Å) | | Cite: | Harnessing the Role of HDAC6 in Idiopathic Pulmonary Fibrosis: Design, Synthesis, Structural Analysis, and Biological Evaluation of Potent Inhibitors.
J.Med.Chem., 64, 2021
|
|
7M1H
 
 | |
7LZP
 
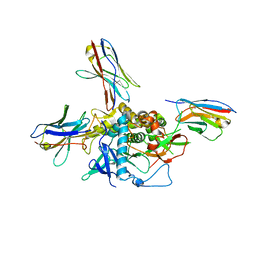 | | LC/A-JPU-B9-JPU-A11-JPU-G11 | | Descriptor: | Botulinum neurotoxin A light chain, JPU-A11, JPU-B9, ... | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2021-03-10 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Probing the structure and function of the protease domain of botulinum neurotoxins using single-domain antibodies.
Plos Pathog., 18, 2022
|
|
6WA2
 
 | | Crystal structure of EGFR(T790M/V948R) in complex with LN3753 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-(3-{5-[2-(acetylamino)pyridin-4-yl]-2-(methylsulfanyl)-1H-imidazol-4-yl}phenyl)-2-fluoro-5-hydroxybenzamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of a "Two-in-One" Mutant-Selective Epidermal Growth Factor Receptor Inhibitor That Spans the Orthosteric and Allosteric Sites.
J.Med.Chem., 65, 2022
|
|
6WAK
 
 | | A crystal structure of EGFR(T790M/V948R) in complex with LN3754 | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, N-(3-{5-[2-(acetylamino)pyridin-4-yl]-2-(methylsulfanyl)-1H-imidazol-4-yl}phenyl)-2-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]benzamide, ... | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of a "Two-in-One" Mutant-Selective Epidermal Growth Factor Receptor Inhibitor That Spans the Orthosteric and Allosteric Sites.
J.Med.Chem., 65, 2022
|
|
4KNE
 
 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in complex with cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Dihydrofolate reductase | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
4KN4
 
 | |
7FH2
 
 | | Crystal structure of the first bromodomain of BRD4 in complex with 16D10 | | Descriptor: | Bromodomain-containing protein 4, CHLORIDE ION, N-[2-[2-[(E)-3-(2,5-dimethoxyphenyl)prop-2-enoyl]-4,5-dimethoxy-phenyl]ethyl]ethanamide | | Authors: | Yokoyama, T, Hirasawa, N. | | Deposit date: | 2021-07-29 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | A chalcone derivative suppresses TSLP induction in mice and human keratinocytes through binding to BET family proteins.
Biochem Pharmacol, 194, 2021
|
|
7EWH
 
 | | Crystal structure of human PHGDH in complex with Homoharringtonine | | Descriptor: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, D-3-phosphoglycerate dehydrogenase | | Authors: | Hsieh, C.H, Cheng, Y.S, Lee, Y.S, Huang, H.C, Juan, H.F. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Homoharringtonine as a PHGDH inhibitor: Unraveling metabolic dependencies and developing a potent therapeutic strategy for high-risk neuroblastoma.
Biomed Pharmacother, 166, 2023
|
|
7L6V
 
 | |
5H0Q
 
 | | Crystal structure of lipid binding protein Nakanori at 1.5A | | Descriptor: | Lipid binding protein | | Authors: | Makino, A, Abe, M, Ishitsuka, R, Murate, M, Kishimoto, T, Sakai, S, Hullin-Matsuda, F, Shimada, Y, Inaba, T, Miyatake, H, Tanaka, H, Kurahashi, A, Pack, C.G, Kasai, R.S, Kubo, S, Schieber, N.L, Dohmae, N, Tochio, N, Hagiwara, K, Sasaki, Y, Aida, Y, Fujimori, F, Kigawa, T, Nishikori, K, Parton, R.G, Kusumi, A, Sako, Y, Anderluh, G, Yamashita, M, Kobayashi, T, Greimel, P, Kobayashi, T. | | Deposit date: | 2016-10-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A novel sphingomyelin/cholesterol domain-specific probe reveals the dynamics of the membrane domains during virus release and in Niemann-Pick type C
FASEB J., 31, 2017
|
|
2ASP
 
 | | Structure of Rabbit Actin In Complex With Reidispongiolide C | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Allingham, J.S, Zampella, A, D'Auria, M.V, Rayment, I. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of microfilament destabilizing toxins bound to actin provide insight into toxin design and activity
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AZB
 
 | | HIV-1 Protease NL4-3 3X mutant in complex with inhibitor, TL-3 | | Descriptor: | PROTEASE RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Kutilek, V, Morris, G.M, Lin, Y.-C, Elder, J.H, Torbett, B.E, Stout, C.D. | | Deposit date: | 2005-09-09 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3
J.Mol.Biol., 356, 2006
|
|
8BSE
 
 | | CRYSTAL STRUCTURE OF SARS-COV-2 RECEPTOR BINDING DOMAIN (RBD) in complex with 1D1 Fab | | Descriptor: | 1D1 FAB HEAVY CHAIN, 1D1 FAB LIGHT CHAIN, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Welin, M, Kimbung, Y.R, Focht, D, Pisitkun, T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Efficacy of the combination of monoclonal antibodies against the SARS-CoV-2 Beta and Delta variants.
Plos One, 18, 2023
|
|
8BSF
 
 | | CRYSTAL STRUCTURE OF SARS-COV-2 RECEPTOR BINDING DOMAIN (RBD-beta variant) in complex with 3D2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3D2 FAB HEAVY CHAIN, 3D2 FAB LIGHT CHAIN, ... | | Authors: | Welin, M, Kimbung, Y.R, Focht, D, Pisitkun, T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Efficacy of the combination of monoclonal antibodies against the SARS-CoV-2 Beta and Delta variants.
Plos One, 18, 2023
|
|
2ASO
 
 | | Structure of Rabbit Actin In Complex With Sphinxolide B | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Allingham, J.S, Zampella, A, D'Auria, M.V, Rayment, I. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of microfilament destabilizing toxins bound to actin provide insight into toxin design and activity
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AZ9
 
 | | HIV-1 Protease NL4-3 1X mutant | | Descriptor: | PROTEASE RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Kutilek, V, Morris, G.M, Lin, Y.-C, Elder, J.H, Torbett, B.E, Stout, C.D. | | Deposit date: | 2005-09-09 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3
J.Mol.Biol., 356, 2006
|
|
8BTS
 
 | |
8B78
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(4~{a}~{R})-8-(2-chloranyl-6-oxidanyl-phenyl)-7-fluoranyl-9-prop-1-ynyl-1,2,4,4~{a},5,11-hexahydropyrazino[2,1-c][1,4]benzoxazepin-3-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C, Breed, J. | | Deposit date: | 2022-09-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Discovery of AZD4747, a Potent and Selective Inhibitor of Mutant GTPase KRAS G12C with Demonstrable CNS Penetration.
J.Med.Chem., 66, 2023
|
|
8B6I
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(4~{a}~{S})-7-chloranyl-8-(5-methyl-2~{H}-indazol-4-yl)-1,2,4,4~{a},5,11-hexahydropyrazino[2,1-c][1,4]benzoxazepin-3-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C, Breed, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of AZD4747, a Potent and Selective Inhibitor of Mutant GTPase KRAS G12C with Demonstrable CNS Penetration.
J.Med.Chem., 66, 2023
|
|
8BHF
 
 | | Cryo-EM structure of stalled rabbit 80S ribosomes in complex with human CCR4-NOT and CNOT4 | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S11, ... | | Authors: | Absmeier, E, Chandrasekaran, V, O'Reilly, F.J, Stowell, J.A.W, Rappsilber, J, Passmore, L.A. | | Deposit date: | 2022-10-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6E53
 
 | | Structure of TERT in complex with a novel telomerase inhibitor | | Descriptor: | MAGNESIUM ION, RNA/DNA hairpin, Telomerase reverse transcriptase, ... | | Authors: | Hernandez-Sanchez, W, Huang, W, Plucinsky, B, Garcia-Vazquez, N, Berdis, A.J, Skordalakes, E, Taylor, D.J. | | Deposit date: | 2018-07-19 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A non-natural nucleotide uses a specific pocket to selectively inhibit telomerase activity.
Plos Biol., 17, 2019
|
|
2AZ8
 
 | | HIV-1 Protease NL4-3 in complex with inhibitor, TL-3 | | Descriptor: | PROTEASE RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Kutilek, V, Morris, G.M, Lin, Y.-C, Elder, J.H, Torbett, B.E, Stout, C.D. | | Deposit date: | 2005-09-09 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3
J.Mol.Biol., 356, 2006
|
|
5UAG
 
 | | Escherichia coli RNA polymerase mutant - RpoB D516V | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.399 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
7KMG
 
 | | LY-CoV555 neutralizing antibody against SARS-CoV-2 | | Descriptor: | GLYCEROL, LY-CoV555 Fab heavy chain, LY-CoV555 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
