7ZH4
 
 | | USP1 bound to ML323 and ubiquitin conjugated to FANCD2 (focused refinement) | | Descriptor: | 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phenyl]methyl]pyrimidin-4-amine, Ubiquitin carboxyl-terminal hydrolase 1, Ubiquitin-60S ribosomal protein L40, ... | | Authors: | Rennie, M.L, Walden, H. | | Deposit date: | 2022-04-05 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site.
Sci Adv, 8, 2022
|
|
8A9J
 
 | |
8A9K
 
 | | Cryo-EM structure of USP1-UAF1 bound to FANCI and mono-ubiquitinated FANCD2 with ML323 (consensus reconstruction) | | Descriptor: | 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phenyl]methyl]pyrimidin-4-amine, DNA (61-MER), Fanconi anemia group D2 protein, ... | | Authors: | Rennie, M.L, Walden, H. | | Deposit date: | 2022-06-28 | | Release date: | 2022-10-12 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site.
Sci Adv, 8, 2022
|
|
7SVS
 
 | |
5K19
 
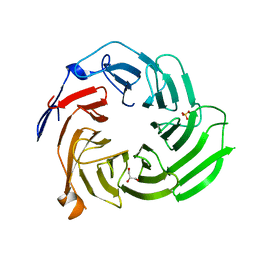 | |
6H4K
 
 | | Structure of the Usp25 C-terminal domain | | Descriptor: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
6HEM
 
 | |
6QP4
 
 | | Structure of 299-452 fragment of the UspA1 protein from Moraxella catarrhalis | | Descriptor: | CHLORIDE ION, HEXANE-1,6-DIOL, UspA1, ... | | Authors: | Mikula, K.M, Kolodziejczyk, R, Goldman, A. | | Deposit date: | 2019-02-13 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the UspA1 protein fragment from Moraxella catarrhalis responsible for C3d binding.
J.Struct.Biol., 208, 2019
|
|
7CJM
 
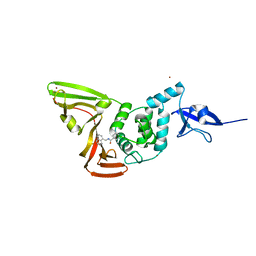 | | SARS CoV-2 PLpro in complex with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Fu, Z, Huang, H. | | Deposit date: | 2020-07-11 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery.
Nat Commun, 12, 2021
|
|
4WY2
 
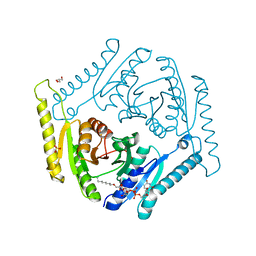 | | Crystal structure of universal stress protein E from Proteus mirabilis in complex with UDP-3-O-[(3R)-3-hydroxytetradecanoyl]-N-acetyl-alpha-glucosamine | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Shumilin, I.A, Shabalin, I.G, Handing, K.B, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-15 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of universal stress protein E from Proteus mirabilis incomplex withUDP-3-O-[(3R)-3-hydroxytetradecanoyl]-N-acetyl-alpha-glucosamine
to be published
|
|
7OWC
 
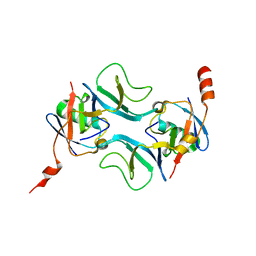 | |
7OWD
 
 | |
2Z84
 
 | | Insights from crystal and solution structures of mouse UfSP1 | | Descriptor: | Ufm1-specific protease 1 | | Authors: | Ha, B.H, Ahn, H.C, Kang, S.H, Tanaka, K, Chung, C.H, Kim, E.E. | | Deposit date: | 2007-08-30 | | Release date: | 2008-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for Ufm1 processing by UfSP1
J. Biol. Chem., 283, 2008
|
|
7L97
 
 | | Crystal structure of STAMBPL1 in complex with an engineered binder | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, SULFATE ION, ... | | Authors: | Guo, Y, Dong, A, Hou, F, Li, Y, Zhang, W, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-01-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional characterization of ubiquitin variant inhibitors for the JAMM-family deubiquitinases STAMBP and STAMBPL1.
J.Biol.Chem., 297, 2021
|
|
1WH0
 
 | | Solution structure of the CS domain of human USP19 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 19 | | Authors: | Nakanishi, T, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CS domain of human USP19
To be Published
|
|
7MS7
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (5-((4-(4-chlorophenyl)piperidin-1-yl)sulfonyl)picolinoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, N-{5-[4-(4-chlorophenyl)piperidine-1-sulfonyl]pyridine-2-carbonyl}glycine, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7MS5
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 4-(4-(4-(3,4-difluoro-phenyl)-piperidin-1-ylsulfonyl)-phenyl)-4-oxo-butanoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[4-(3,4-difluorophenyl)piperidine-1-sulfonyl]phenyl}-4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7MS6
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (2-fluoro-4-((4-phenylpiperidin-1-yl)sulfonyl)benzoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, N-[2-fluoro-4-(4-phenylpiperidine-1-sulfonyl)benzoyl]glycine, SULFATE ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
8FWP
 
 | | Crystal Structure of CDC10 - CDC3 heterocomplex from Saccharomyces cerevisiae | | Descriptor: | Cell division control protein 10, Cell division control protein 3, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Silva, R.M, Leonardo, D.A, Pereira, H.M, Garratt, R.C. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A key piece of the puzzle: The central tetramer of the Saccharomyces cerevisiae septin protofilament and its implications for self-assembly.
J.Struct.Biol., 215, 2023
|
|
8G6P
 
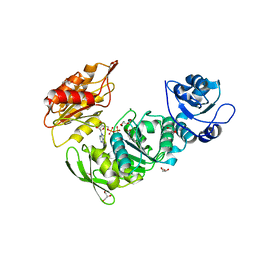 | | Crystal structure of Mycobacterium thermoresistibile MurE in complex with ADP and 2,6-Diaminopimelic acid | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Rossini, N.O, Silva, C.S, Dias, M.V.B. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of Mycobacterium thermoresistibile MurE ligase reveals the binding mode of the substrate m-diaminopimelate.
J.Struct.Biol., 215, 2023
|
|
6BCM
 
 | |
8GF3
 
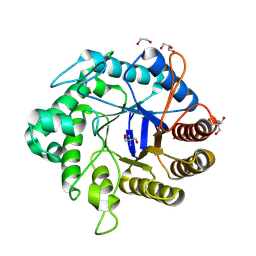 | | Crystallographic structure from BlMan5_7 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Briganti, L, Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-03-07 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic structure from BlMan5_7
To be published
|
|
2DAG
 
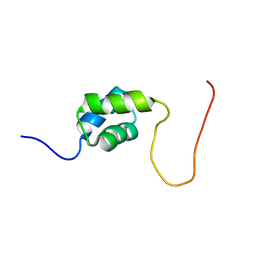 | | Solution Structure of the First UBA Domain in the Human Ubiquitin Specific Protease 5 (Isopeptidase 5) | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 5 | | Authors: | Zhao, C, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-06-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the First UBA Domain in the Human Ubiquitin Specific Protease 5 (Isopeptidase 5)
To be Published
|
|
2DAK
 
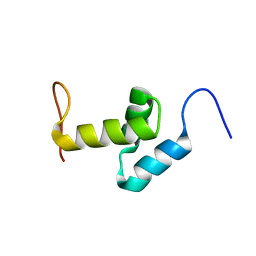 | | Solution Structure of the Second UBA Domain in the Human Ubiquitin Specific Protease 5 (Isopeptidase 5) | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 5 | | Authors: | Zhao, C, Kigawa, T, Sato, M, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-06-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Second UBA Domain in the Human Ubiquitin Specific Protease 5 (Isopeptidase 5)
To be Published
|
|
5V5T
 
 | | Crystal structure of leucine-rich protein regulator, ElrR, from Enterococcus faecalis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Conserved domain protein | | Authors: | De Groote, M.C.R, Camargo, I.L, Serror, P, Horjales, E. | | Deposit date: | 2017-03-15 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure and functional characterization of ElrR, a member of the RRNPP family of transcriptional regulators.
To be published
|
|
