1T09
 
 | | Crystal structure of human cytosolic NADP(+)-dependent isocitrate dehydrogenase in complex NADP | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Xu, X, Zhao, J, Peng, B, Huang, Q, Arnold, E, Ding, J. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity
J.Biol.Chem., 279, 2004
|
|
1T0A
 
 | | Crystal Structure of 2C-Methyl-D-Erythritol-2,4-cyclodiphosphate Synthase from Shewanella Oneidensis | | Descriptor: | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, COBALT (II) ION, FARNESYL DIPHOSPHATE, ... | | Authors: | Ni, S, Robinson, H, Marsing, G.C, Bussiere, D.E, Kennedy, M.A. | | Deposit date: | 2004-04-08 | | Release date: | 2004-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase from Shewanella oneidensis at 1.6 A: identification of farnesyl pyrophosphate trapped in a hydrophobic cavity.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1T0B
 
 | | Structure of ThuA-like protein from Bacillus stearothermophilus | | Descriptor: | ThuA-like protein, ZINC ION | | Authors: | Borek, D, Chen, Y, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-04-08 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of chemical interactions in ThuA-like protein from Bacillus Stearothermophilus
To be Published
|
|
1T0C
 
 | |
1T0D
 
 | | Crystal Structure of 2-aminopurine labelled bacterial decoding site RNA | | Descriptor: | 5'-R(*CP*AP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*CP*C)-3', 5'-R(*GP*GP*UP*GP*GP*UP*GP*(MTU)P*AP*GP*UP*CP*GP*CP*UP*GP*G)-3' | | Authors: | Shandrick, S, Zhao, Q, Han, Q, Ayida, B.K, Takahashi, M, Winters, G.C, Simonsen, K.B, Vourloumis, D, Hermann, T. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Monitoring molecular recognition of the ribosomal decoding site.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
1T0E
 
 | | Crystal Structure of 2-aminopurine labelled bacterial decoding site RNA | | Descriptor: | 5'-R(*CP*GP*AP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*CP*C)-3', 5'-R(*GP*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*UP*CP*GP*G)-3', SULFATE ION | | Authors: | Shandrick, S, Zhao, Q, Han, Q, Ayida, B.K, Takahashi, M, Winters, G.C, Simonsen, K.B, Vourloumis, D, Hermann, T. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Monitoring molecular recognition of the ribosomal decoding site.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
1T0F
 
 | | Crystal Structure of the TnsA/TnsC(504-555) complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, MALONIC ACID, ... | | Authors: | Ronning, D.R, Li, Y, Perez, Z.N, Ross, P.D, Hickman, A.B, Craig, N.L, Dyda, F. | | Deposit date: | 2004-04-08 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The carboxy-terminal portion of TnsC activates the Tn7 transposase through a specific interaction with TnsA.
Embo J., 23, 2004
|
|
1T0G
 
 | | Hypothetical protein At2g24940.1 from Arabidopsis thaliana has a cytochrome b5 like fold | | Descriptor: | cytochrome b5 domain-containing protein | | Authors: | Song, J, Vinarov, D.A, Tyler, E.M, Shahan, M.N, Tyler, R.C, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-04-08 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hypothetical protein At2g24940.1 from Arabidopsis thaliana has a cytochrome b5 like fold
J.Biomol.NMR, 30, 2004
|
|
1T0H
 
 | | Crystal structure of the Rattus norvegicus voltage gated calcium channel beta subunit isoform 2a | | Descriptor: | CHLORIDE ION, VOLTAGE-GATED CALCIUM CHANNEL SUBUNIT BETA2A | | Authors: | Van Petegem, F, Clark, K, Chatelain, F, Minor Jr, D. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of a complex between a voltage-gated calcium channel beta-subunit and an alpha-subunit domain.
Nature, 429, 2004
|
|
1T0I
 
 | | YLR011wp, a Saccharomyces cerevisiae NA(D)PH-dependent FMN reductase | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, YLR011wp | | Authors: | Liger, D, Graille, M, Zhou, C.-Z, Leulliot, N, Quevillon-Cheruel, S, Blondeau, K, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2004-04-09 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Characterization of Yeast YLR011wp, an Enzyme with NAD(P)H-FMN and Ferric Iron Reductase Activities
J.Biol.Chem., 279, 2004
|
|
1T0J
 
 | | Crystal structure of a complex between voltage-gated calcium channel beta2a subunit and a peptide of the alpha1c subunit | | Descriptor: | CHLORIDE ION, Voltage-dependent L-type calcium channel alpha-1C subunit, voltage-gated calcium channel subunit beta2a | | Authors: | Van Petegem, F, Clark, K, Chatelain, F, Minor Jr, D. | | Deposit date: | 2004-04-09 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a complex between a voltage-gated calcium channel beta-subunit and an alpha-subunit domain.
Nature, 429, 2004
|
|
1T0K
 
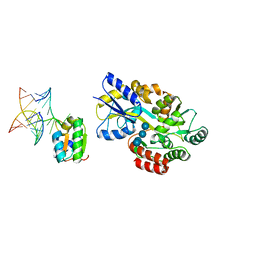 | | Joint X-ray and NMR Refinement of Yeast L30e-mRNA complex | | Descriptor: | 5'-R(*G*GP*AP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*GP*UP*C)-3', 5'-R(*GP*AP*CP*CP*GP*GP*AP*GP*UP*GP*UP*CP*C)-3', 60S ribosomal protein L30, ... | | Authors: | Chao, J.A, Williamson, J.R. | | Deposit date: | 2004-04-09 | | Release date: | 2004-07-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Joint X-Ray and NMR Refinement of the Yeast L30e-mRNA Complex
Structure, 12, 2004
|
|
1T0L
 
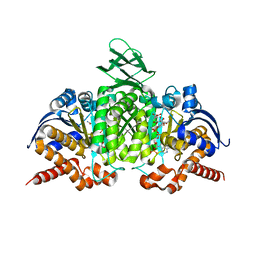 | | Crystal structure of human cytosolic NADP(+)-dependent isocitrate dehydrogenase in complex with NADP, isocitrate, and calcium(2+) | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Xu, X, Zhao, J, Peng, B, Huang, Q, Arnold, E, Ding, J. | | Deposit date: | 2004-04-10 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity
J.Biol.Chem., 279, 2004
|
|
1T0M
 
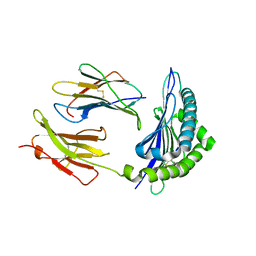 | | Conformational switch in polymorphic H-2K molecules containing an HSV peptide | | Descriptor: | Beta-2-microglobulin, Glycoprotein B, H-2 class I histocompatibility antigen, ... | | Authors: | Webb, A.I, Borg, N.A, Dunstone, M.A, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Carbone, F.R, Bottomley, S.P, Purcell, A.W, Rossjohn, J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-11-23 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of H-2K(b) and K(bm8) complexed to a herpes simplex virus determinant: evidence for a conformational switch that governs T cell repertoire selection and viral resistance.
J Immunol., 173, 2004
|
|
1T0N
 
 | | Conformational switch in polymorphic H-2K molecules containing an HSV peptide | | Descriptor: | Beta-2-microglobulin, Glycoprotein B, H-2 class I histocompatibility antigen, ... | | Authors: | Webb, A.I, Borg, N.A, Dunstone, M.A, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Carbone, F.R, Bottomley, S.P, Purcell, A.W, Rossjohn, J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-11-23 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of H-2K(b) and K(bm8) complexed to a herpes simplex virus determinant: evidence for a conformational switch that governs T cell repertoire selection and viral resistance.
J Immunol., 173, 2004
|
|
1T0O
 
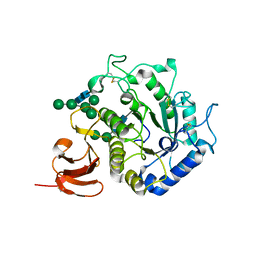 | | The structure of alpha-galactosidase from Trichoderma reesei complexed with beta-D-galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Golubev, A.M, Nagem, R.A.P, Brandao Neto, J.R, Neustroev, K.N, Eneyskaya, E.V, Kulminskaya, A.A, Shabalin, K.A, Savel'ev, A.N, Polikarpov, I. | | Deposit date: | 2004-04-12 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of alpha-galactosidase from Trichoderma reesei and its complex with galactose: implications for catalytic mechanism
J.Mol.Biol., 339, 2004
|
|
1T0P
 
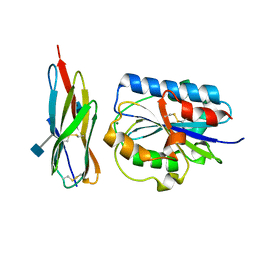 | | Structural Basis of ICAM recognition by integrin alpahLbeta2 revealed in the complex structure of binding domains of ICAM-3 and alphaLbeta2 at 1.65 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-L, Intercellular adhesion molecule-3, ... | | Authors: | Song, G, Yang, Y.T, Liu, J.H, Shimaoko, M, Springer, T.A, Wang, J.H. | | Deposit date: | 2004-04-12 | | Release date: | 2005-03-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An atomic resolution view of ICAM recognition in a complex between the binding domains of ICAM-3 and integrin alphaLbeta2.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1T0Q
 
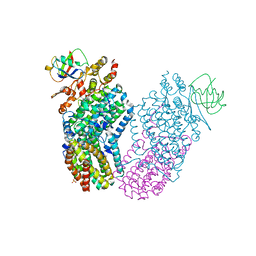 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase | | Descriptor: | FE (III) ION, HYDROXIDE ION, SULFANYLACETIC ACID, ... | | Authors: | Sazinsky, M.H, Bard, J, Di Donato, A, Lippard, S.J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase from Pseudomonas stutzeri OX1: INSIGHT INTO THE SUBSTRATE SPECIFICITY, SUBSTRATE CHANNELING, AND ACTIVE SITE TUNING OF MULTICOMPONENT MONOOXYGENASES.
J.Biol.Chem., 279, 2004
|
|
1T0R
 
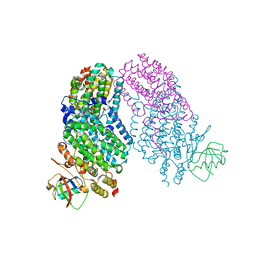 | | Crystal Structure of the Toluene/o-xylene Monooxygenase Hydroxuylase from Pseudomonas stutzeri-azide bound | | Descriptor: | AZIDE ION, FE (III) ION, HYDROXIDE ION, ... | | Authors: | Sazinsky, M.H, Bard, J, Di Donato, A, Lippard, S.J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase from Pseudomonas stutzeri OX1: INSIGHT INTO THE SUBSTRATE SPECIFICITY, SUBSTRATE CHANNELING, AND ACTIVE SITE TUNING OF MULTICOMPONENT MONOOXYGENASES.
J.Biol.Chem., 279, 2004
|
|
1T0S
 
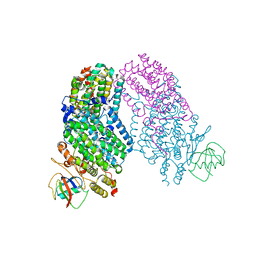 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase with 4-bromophenol bound | | Descriptor: | 4-BROMOPHENOL, FE (III) ION, HYDROXIDE ION, ... | | Authors: | Sazinsky, M.H, Bard, J, Di Donato, A, Lippard, S.J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase from Pseudomonas stutzeri OX1: INSIGHT INTO THE SUBSTRATE SPECIFICITY, SUBSTRATE CHANNELING, AND ACTIVE SITE TUNING OF MULTICOMPONENT MONOOXYGENASES.
J.Biol.Chem., 279, 2004
|
|
1T0T
 
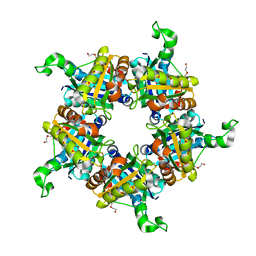 | | Crystallographic structure of a putative chlorite dismutase | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, APC35880, MAGNESIUM ION | | Authors: | Gilski, M, Borek, D, Chen, Y, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-04-12 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of APC35880 protein from Bacillus Stearothermophilus
To be Published
|
|
1T0U
 
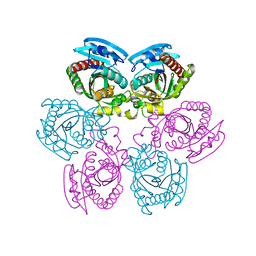 | | Crystal structure of E.coli uridine phosphorylase at 2.2 A resolution (Type-A Native) | | Descriptor: | Uridine phosphorylase | | Authors: | Caradoc-Davies, T.T, Cutfield, S.M, Lamont, I.L, Cutfield, J.F. | | Deposit date: | 2004-04-13 | | Release date: | 2004-04-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of escherichia coli uridine phosphorylase in two native and three complexed forms reveal basis of substrate specificity, induced conformational changes and influence of potassium
J.Mol.Biol., 337, 2004
|
|
1T0V
 
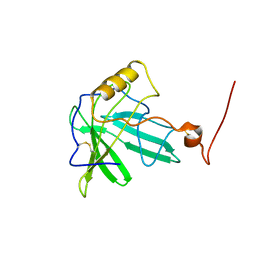 | | NMR Solution Structure of the Engineered Lipocalin FluA(R95K) Northeast Structural Genomics Target OR17 | | Descriptor: | BILIN-BINDING PROTEIN | | Authors: | Mills, J.L, Liu, G, Skerra, A, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-04-13 | | Release date: | 2005-06-14 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of the engineered fluorescein-binding lipocalin FluA reveal rigidification of beta-barrel and variable loops upon enthalpy-driven ligand binding.
Biochemistry, 48, 2009
|
|
1T0W
 
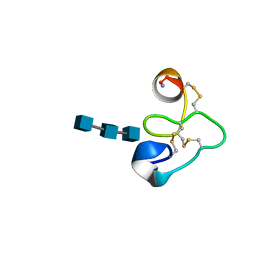 | | 25 NMR structures of Truncated Hevein of 32 aa (Hevein-32) complex with N,N,N-triacetylglucosamina | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevein | | Authors: | Aboitiz, N, Vila-Perello, M, Groves, P, Asensio, J.L, Andreu, D, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2004-04-13 | | Release date: | 2004-09-28 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | NMR and modeling studies of protein-carbohydrate interactions: synthesis, three-dimensional structure, and recognition properties of a minimum hevein domain with binding affinity for chitooligosaccharides
Chembiochem, 5, 2004
|
|
1T0Y
 
 | | Solution Structure of a Ubiquitin-Like Domain from Tubulin-binding Cofactor B | | Descriptor: | tubulin folding cofactor B | | Authors: | Lytle, B.L, Peterson, F.C, Qui, S.H, Luo, M, Volkman, B.F, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Ubiquitin-like Domain from Tubulin-binding Cofactor B.
J.Biol.Chem., 279, 2004
|
|
