9QGB
 
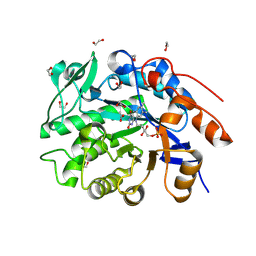 | | Crystal structure of an NADH-accepting ene reductase variant NostocER1-L1,5 (engineered Loop Swap from Achromobacter sp. JA81) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, All1865 protein, ... | | Authors: | Bischoff, D, Walla, B, Janowski, R, Maslakova, A, Maehler, C, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2025-03-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Rational Introduction of Electrostatic Interactions at Crystal Contacts to Enhance Protein Crystallization of an Ene Reductase.
Biomolecules, 15, 2025
|
|
9QM8
 
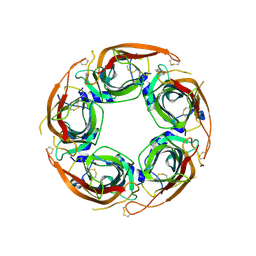 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with IOTA739 | | Descriptor: | 1,10-PHENANTHROLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Cederfelt, D, Lund, B.A, Boronat, P, Hennig, S, Dobritzsch, D, Danielson, U.H. | | Deposit date: | 2025-03-22 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
To Be Published
|
|
4W2H
 
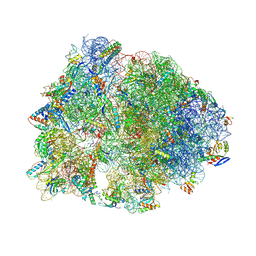 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with pactamycin (co-crystallized), mRNA and deacylated tRNA in the P site | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Osterman, I.A, Szal, T, Tashlitsky, V.N, Serebryakova, M.V, Kusochek, P, Bulkley, D, Malanicheva, I.A, Efimenko, T.A, Efremenkova, O.V, Konevega, A.L, Shaw, K.J, Bogdanov, A.A, Rodnina, M.V, Dontsova, O.A, Mankin, A.S, Steitz, T.A, Sergiev, P.V. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome.
Mol.Cell, 56, 2014
|
|
4V4P
 
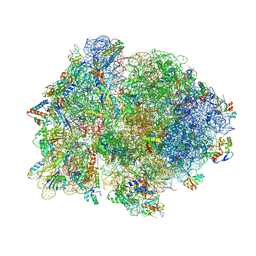 | | Crystal structure of 70S ribosome with thrS operator and tRNAs. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Romby, P, Rees, B, Schulze-Briese, C, Springer, M, Ehresmann, C, Ehresmann, B, Moras, D, Yusupova, G, Yusupov, M. | | Deposit date: | 2005-01-19 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Translational operator of mRNA on the ribosome: how repressor proteins exclude ribosome binding.
Science, 308, 2005
|
|
4V4E
 
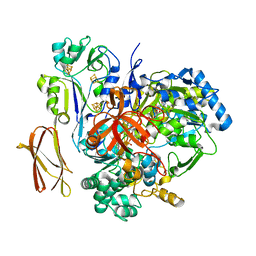 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici complexed with inhibitor 1,2,4,5-tetrahydroxy-benzene | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, BENZENE-1,2,4,5-TETROL, CALCIUM ION, ... | | Authors: | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | Deposit date: | 2004-06-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
4W9A
 
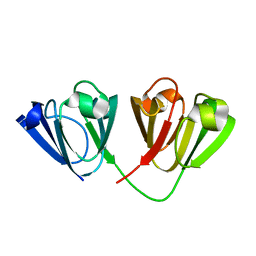 | | Crystal structure of Gamma-B Crystallin expressed in E. coli based on mRNA variant 2 | | Descriptor: | Gamma-crystallin B | | Authors: | Kudlinzki, D, Buhr, F, Linhard, V.L, Jha, S, Komar, A.A, Schwalbe, H. | | Deposit date: | 2014-08-27 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Two synonymous gene variants encode proteins with identical sequence, but different folding conformations.
To Be Published
|
|
4V1N
 
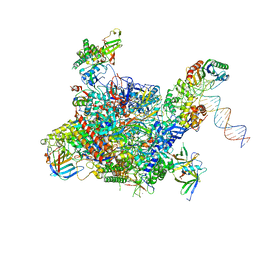 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | Descriptor: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7A1U
 
 | | Structure of SARS-CoV-2 Main Protease bound to Fusidic Acid. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FUSIDIC ACID, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
2X5O
 
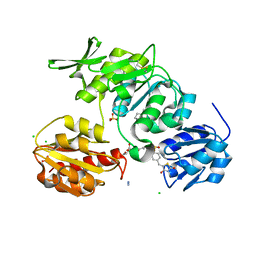 | | Discovery of Novel 5-Benzylidenerhodanine- and 5-Benzylidene- thiazolidine-2,4-dione Inhibitors of MurD Ligase | | Descriptor: | AZIDE ION, CHLORIDE ION, N-({3-[({4-[(Z)-(2,4-DIOXO-1,3-THIAZOLIDIN-5-YLIDENE)METHYL]PHENYL}AMINO)METHYL]PHENYL}CARBONYL)-D-GLUTAMIC ACID, ... | | Authors: | Zidar, N, Tomasic, T, Sink, R, Rupnik, V, Kovac, A, Turk, S, Contreras-Martel, C, Dessen, A, Blanot, D, Gobec, S, Zega, A, Peterlin-Masic, L, Kikelja, D. | | Deposit date: | 2010-02-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of novel 5-benzylidenerhodanine and 5-benzylidenethiazolidine-2,4-dione inhibitors of MurD ligase.
J. Med. Chem., 53, 2010
|
|
2WPA
 
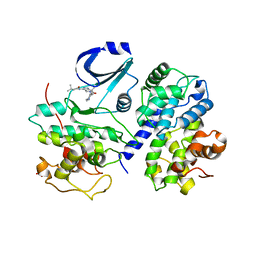 | | Optimisation of 6,6-Dimethyl Pyrrolo 3,4-c pyrazoles: Identification of PHA-793887, a Potent CDK Inhibitor Suitable for Intravenous Dosing | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, N-{6,6-DIMETHYL-5-[(1-METHYLPIPERIDIN-4-YL)CARBONYL]-1,4,5,6-TETRAHYDROPYRROLO[3,4-C]PYRAZOL-3-YL}-3-METHYLBUTANAMIDE, ... | | Authors: | Brasca, M.G, Albanese, C, Alzani, R, Amici, R, Avanzi, N, Ballinari, D, Bischoff, J, Borghi, D, Casale, E, Croci, V, Fiorentini, F, Isacchi, A, Mercurio, C, Nesi, M, Orsini, P, Pastori, W, Pesenti, E, Pevarello, P, Roussel, P, Varasi, M, Volpi, D, Vulpetti, A, Ciomei, M. | | Deposit date: | 2009-08-03 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Optimization of 6,6-Dimethyl Pyrrolo[3,4-C]Pyrazoles: Identification of Pha-793887, a Potent Cdk Inhibitor Suitable for Intravenous Dosing.
Bioorg.Med.Chem., 18, 2010
|
|
2WJP
 
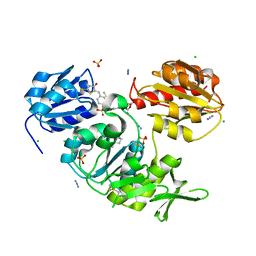 | | CRYSTAL STRUCTURE OF MURD LIGASE IN COMPLEX WITH D-GLU CONTAINING RHODANINE INHIBITOR | | Descriptor: | AZIDE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Tomasic, T, Zidar, N, Sink, R, Kovac, A, Rupnik, V, Turk, S, Contreras-Martel, C, Dessen, A, Blanot, D, Muller-Premru, M, Gobec, S, Zega, A, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2009-05-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Novel 5-Benzylidenerhodanine and 5-Benzylidenethiazolidine-2,4-Dione Inhibitors of Murd Ligase.
J.Med.Chem., 53, 2010
|
|
1RNQ
 
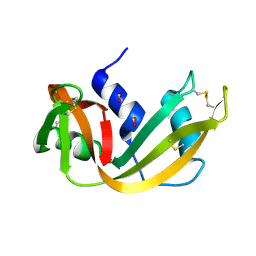 | | RIBONUCLEASE A CRYSTALLIZED FROM 8M SODIUM FORMATE | | Descriptor: | FORMIC ACID, RIBONUCLEASE A | | Authors: | Fedorov, A.A, Josef-Mccarthy, D, Graf, I, Anguelova, D, Fedorov, E.V, Almo, S.C. | | Deposit date: | 1995-11-08 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ionic interactions in crystalline bovine pancreatic ribonuclease A.
Biochemistry, 35, 1996
|
|
6OOZ
 
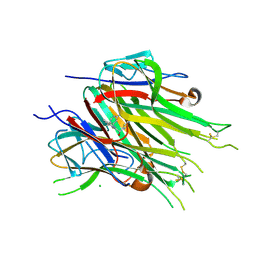 | | Asymmetric hTNF-alpha | | Descriptor: | (S)-{1-[(2,5-dimethylphenyl)methyl]-1H-benzimidazol-2-yl}(pyridin-4-yl)methanol, CHLORIDE ION, Tumor necrosis factor | | Authors: | Arakaki, T.L, Edwards, T.E, Fox III, D, Lecomte, F, Ceska, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Small molecules that inhibit TNF signalling by stabilising an asymmetric form of the trimer.
Nat Commun, 10, 2019
|
|
1R0Q
 
 | | Characterization of the conversion of the malformed, recombinant cytochrome rc552 to a 2-formyl-4-vinyl (Spirographis) heme | | Descriptor: | 2-FORMYL-PROTOPORPHRYN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-22 | | Release date: | 2004-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
1QYZ
 
 | | Characterization of the malformed, recombinant cytochrome rC552 | | Descriptor: | 2-ACETYL-PROTOPORPHYRIN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
9QWO
 
 | | Vinculin tail bound to paxillin LD2 | | Descriptor: | ACETATE ION, Isoform 1 of Vinculin, Isoform Gamma of Paxillin, ... | | Authors: | Diaz-Palacios, K, Lietha, D. | | Deposit date: | 2025-04-14 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Phospho-regulated tethering of focal adhesion kinase to vinculin links force transduction to focal adhesion signaling.
Cell Commun Signal, 23, 2025
|
|
9UBG
 
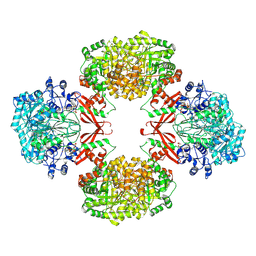 | | Cryo-EM structure of Pyruvate carboxylase from Mycobacterium tuberculosis in complex with Acetyl-CoA and ADP | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, Pyruvate carboxylase, ... | | Authors: | Singh, A, Sharma, D, Raza, M, Singh, S, Das, U. | | Deposit date: | 2025-04-03 | | Release date: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of Pyruvate carboxylase from Mycobacterium tuberculosis in complex with Acetyl-CoA and ADP
To Be Published
|
|
9NYI
 
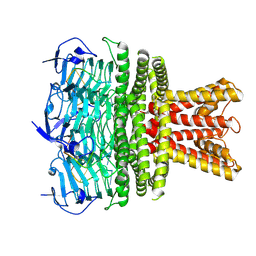 | | Structure of HalA in complex with oligodeoxyadenylate | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*A)-3'), Structure of HalA in complex with oligodeoxyadenylate | | Authors: | Tan, J.M.J, Melamed, S, Cofsky, J.C, Syangtan, D, Hobbs, S.J, Del Marmol, J, Jost, M, Kruse, A.C, Sorek, S, Kranzusch, P.J. | | Deposit date: | 2025-03-27 | | Release date: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Structure of Hailong HalA in complex with oligodeoxyadenylate
To Be Published
|
|
9QUW
 
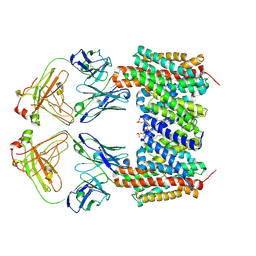 | | Cryo-EM structure of the human NHA2-Fab complex bound to phloretin | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, 3-(4-HYDROXYPHENYL)-1-(2,4,6-TRIHYDROXYPHENYL)PROPAN-1-ONE, Fab heavy chain, ... | | Authors: | Jung, S, Drew, D. | | Deposit date: | 2025-04-11 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and Inhibition of the Human Na + /H + Exchanger SLC9B2.
Int J Mol Sci, 26, 2025
|
|
9UEQ
 
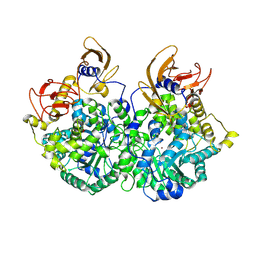 | | Cryo-EM structure of CT-BCCP domain of Pyruvate carboxylase from Mycobacterium tuberculosis | | Descriptor: | BIOTIN, OXALATE ION, Pyruvate carboxylase, ... | | Authors: | Singh, A, Sharma, D, Raza, M, Singh, S, Das, U. | | Deposit date: | 2025-04-09 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of Pyruvate carboxylase from Mycobacterium tuberculosis in complex with Acetyl-CoA and ADP
To Be Published
|
|
9NZH
 
 | |
9UGE
 
 | | Crystal structure of the complex of camel peptidoglycan recognition protein, PGRP-S with malic acid and oxalic acid at 2.3 A resolution | | Descriptor: | D-MALATE, OXALIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Barik, D, Ahmad, N, Maurya, A, Yamini, S, Sharma, P, Yadav, S.P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2025-04-11 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Crystal structure of the complex of camel peptidoglycan recognition protein, PGRP-S with malic acid and oxalic acid at 2.3 A resolution
To Be Published
|
|
9NIU
 
 | | Co-crystal structure of FABP7 in complex with PFO3TDA | | Descriptor: | 1,2-ETHANEDIOL, Fatty acid-binding protein, brain, ... | | Authors: | Yang, D, Liu, J, Zeng, H, Dong, A, Arrowsmith, C.H, Edwards, A.M, Peng, H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-02-26 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Co-crystal structure of FABP7 in complex with PFO3TDA
To be published
|
|
1HOY
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN A-BUNGAROTOXIN AND A MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Descriptor: | LONG NEUROTOXIN 1, MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Authors: | Scarselli, M, Spiga, O, Ciutti, A, Bracci, L, Lelli, B, Lozzi, L, Calamandrei, D, Bernini, A, Di Maro, D, Niccolai, N, Neri, P. | | Deposit date: | 2000-12-12 | | Release date: | 2000-12-27 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
