8CNM
 
 | |
7MAF
 
 | | HIV-1 Protease (I84V) in Complex with PU2 (LR2-79) | | Descriptor: | Protease, SULFATE ION, diethyl ({4-[(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}butyl]phenoxy}methyl)phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with PU2 (LR2-79)
To Be Published
|
|
7MAB
 
 | | HIV-1 Protease (I84V) in Complex with GS-8374 | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with GS-8374
To Be Published
|
|
6VPY
 
 | | I33M (I3.2 mutant from CH103 Lineage) | | Descriptor: | CHLORIDE ION, GLYCEROL, I33M heavy chain, ... | | Authors: | Fera, D, Zhou, J. | | Deposit date: | 2020-02-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The Effects of Framework Mutations at the Variable Domain Interface on Antibody Affinity Maturation in an HIV-1 Broadly Neutralizing Antibody Lineage.
Front Immunol, 11, 2020
|
|
7LLQ
 
 | | Substrate-dependent divergence of leukotriene A4 hydrolase aminopeptidase activity | | Descriptor: | 1-benzyl-4-methoxybenzene, 1-{4-oxo-4-[(2S)-pyrrolidin-2-yl]butanoyl}-L-proline, Leukotriene A-4 hydrolase, ... | | Authors: | Lee, K.H, Lee, S.H, Shim, Y.M, Paige, M, Noble, S.M. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Substrate-dependent modulation of the leukotriene A 4 hydrolase aminopeptidase activity and effect in a murine model of acute lung inflammation.
Sci Rep, 12, 2022
|
|
4R4K
 
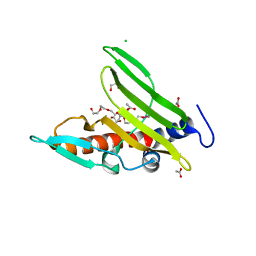 | | Crystal structure of a cystatin-like protein (BACCAC_01506) from Bacteroides caccae ATCC 43185 at 1.69 A resolution | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of a hypothetical protein (BACCAC_01506) from Bacteroides caccae ATCC 43185 at 1.69 A resolution
To be published
|
|
4QSX
 
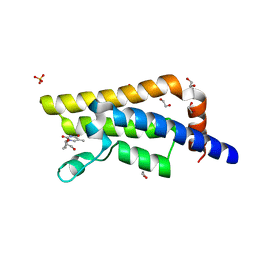 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with 3'-deoxy thymidine | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2R,5S)-5-(hydroxymethyl)tetrahydrofuran-2-yl]-5-methylpyrimidine-2,4(1H,3H)-dione, ATPase family AAA domain-containing protein 2, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
8U8B
 
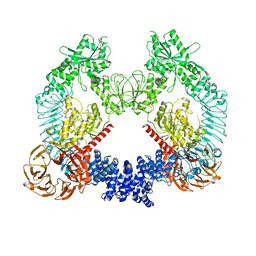 | | Cryo-EM structure of LRRK2 bound to type II inhibitor rebastinib | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-09-16 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM.
Cell Discov, 10, 2024
|
|
4WBE
 
 | |
7P1A
 
 | | Carbonic Anhydrase VII Sultam Based Inhibitors | | Descriptor: | 4-[2-[4-[(4-methylphenyl)methyl]-1,1-bis(oxidanylidene)-1,2,4-thiadiazinan-2-yl]ethyl]benzenesulfonamide, Carbonic anhydrase 7, GLYCEROL, ... | | Authors: | D'Ambrosio, K, De Simone, G. | | Deposit date: | 2021-07-01 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Sultam based Carbonic Anhydrase VII inhibitors for the management of neuropathic pain.
Eur.J.Med.Chem., 227, 2021
|
|
8F29
 
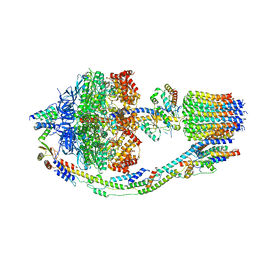 | | Yeast ATP synthase in conformation-1 at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-07 | | Release date: | 2024-02-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6YID
 
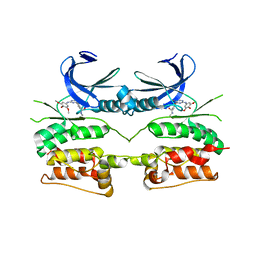 | | Crystal structure of ULK2 in complex with SBI-0206965 | | Descriptor: | 2-({5-bromo-2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}oxy)-N-methylbenzene-1-carboximidic acid, Serine/threonine-protein kinase ULK2 | | Authors: | Chaikuad, A, Ren, H, Bakas, N.A, Lambert, L.J, Cosford, N.D.P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, Synthesis, and Characterization of an Orally Active Dual-Specific ULK1/2 Autophagy Inhibitor that Synergizes with the PARP Inhibitor Olaparib for the Treatment of Triple-Negative Breast Cancer.
J.Med.Chem., 63, 2020
|
|
6W3A
 
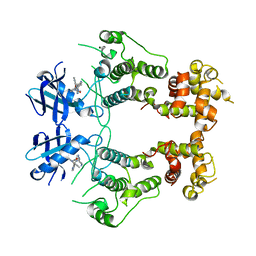 | | Structure of unphosphorylated IRE1 in complex with G-7658 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1, ~{N}-[6-methyl-5-[3-[2-[[(3~{S})-piperidin-3-yl]amino]pyrimidin-4-yl]pyridin-2-yl]oxy-naphthalen-1-yl]but-2-ynamide | | Authors: | Ferri, E, Wang, W, Joachim, R, Mortara, K. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
7OZB
 
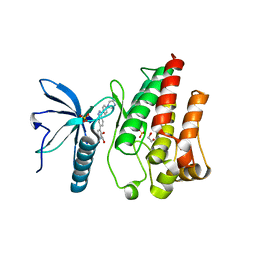 | | FGFR1 kinase domain (residues 458-765) with mutations C488A, C584S in complex with 38. | | Descriptor: | 1,2-ETHANEDIOL, 4-[3-(4-piperazin-4-ium-1-ylphenyl)-1H-indazol-6-yl]phenol, Fibroblast growth factor receptor 1, ... | | Authors: | Trinh, C.H, Turner, L.D, Fishwick, C.W.G. | | Deposit date: | 2021-06-27 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | From Fragment to Lead: De Novo Design and Development toward a Selective FGFR2 Inhibitor.
J.Med.Chem., 65, 2022
|
|
6YG3
 
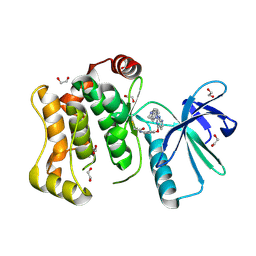 | | Crystal structure of MKK7 (MAP2K7) covalently bound with CPT1-70-1 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 7, N-(4-((2-((4-(4-methylpiperazin-1-yl)phenyl)amino)-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy)phenyl)acrylamide | | Authors: | Chaikuad, A, Tan, L, Wang, J, Liang, Y, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
6OOB
 
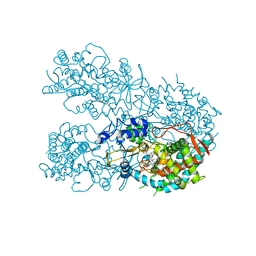 | | Human CYP3A4 bound to a suicide substrate | | Descriptor: | 4-{[(2Z,6S)-6,7-dihydroxy-3,7-dimethyloct-2-en-1-yl]oxy}-7H-furo[3,2-g][1]benzopyran-7-one, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-04-22 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural Insights into the Interaction of Cytochrome P450 3A4 with Suicide Substrates: Mibefradil, Azamulin and 6',7'-Dihydroxybergamottin.
Int J Mol Sci, 20, 2019
|
|
8OZW
 
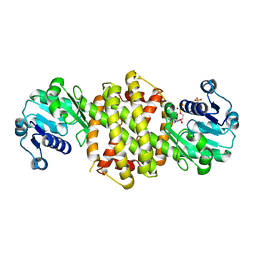 | | Imine Reductase from Ajellomyces dermatitidis in complex NADPH4 | | Descriptor: | 1,2-ETHANEDIOL, 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Sharma, M, Grogan, G. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the imine reductase from Ajellomyces dermatitidis in three crystal forms.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8TS6
 
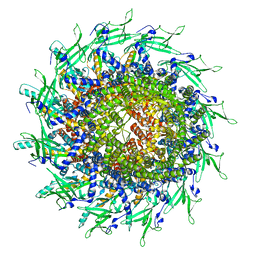 | | Cyanophage A-1(L) portal | | Descriptor: | Portal protein | | Authors: | Yu, R.C, Li, Q, Zhou, C.Z. | | Deposit date: | 2023-08-11 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structure of the intact tail machine of Anabaena myophage A-1(L).
Nat Commun, 15, 2024
|
|
6YPZ
 
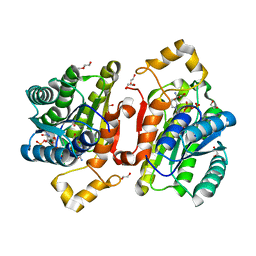 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6W61
 
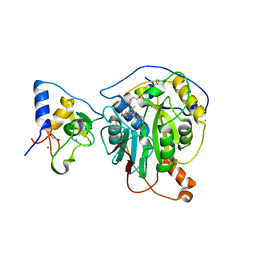 | | Crystal Structure of the methyltransferase-stimulatory factor complex of NSP16 and NSP10 from SARS CoV-2. | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, CHLORIDE ION, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine
Biorxiv, 2020
|
|
6W9I
 
 | |
7MFO
 
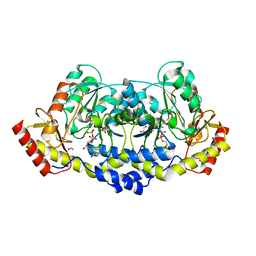 | | X-ray structure of the L136 Aminotransferase from Acanthamoeba polyphaga mimivirus in the presence of TDP and PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CHLORIDE ION, ... | | Authors: | Ferek, J.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-04-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
6YMA
 
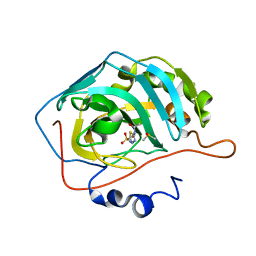 | | MicroED structure of acetazolamide-bound human carbonic anhydrase II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, DIMETHYL SULFOXIDE, ZINC ION, ... | | Authors: | Clabbers, M.T.B, Fisher, S.Z, Coincon, M, Zou, X, Xu, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Visualizing drug binding interactions using microcrystal electron diffraction.
Commun Biol, 3, 2020
|
|
8FQX
 
 | | Carbonic Anhydrase II in complex with the alkyl ureas 3g | | Descriptor: | 4-hydroxy-3-({[(pyridin-4-yl)methyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Combs, J.E, McKenna, R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | New Insights into Conformationally Restricted Carbonic Anhydrase Inhibitors.
Molecules, 28, 2023
|
|
9E6Y
 
 | |
