2HP5
 
 | | Crystal Structure of the OXA-10 W154G mutant at pH 7.0 | | Descriptor: | Beta-lactamase PSE-2, COBALT (II) ION, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2HP6
 
 | | Crystal structure of the OXA-10 W154A mutant at pH 7.5 | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2HP9
 
 | | Crystal Structure of the OXA-10 W154A mutant at pH 6.0 | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2I72
 
 | | AmpC beta-lactamase in complex with 5-diformylaminomethyl-benzo[b]thiophen-2-boronic acid | | Descriptor: | Beta-lactamase, {5-[(DIFORMYLAMINO)METHYL]-1-BENZOTHIEN-2-YL}BORONIC ACID | | Authors: | Venturelli, A, Cancian, L, Tondi, D, Morandi, F, Cannazza, G, Segatore, B, Prati, F, Amicosante, G, Shoichet, B.K, Costi, M.P. | | Deposit date: | 2006-08-30 | | Release date: | 2007-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimizing Cell Permeation of an Antibiotic Resistance Inhibitor for Improved Efficacy
J.Med.Chem., 50, 2007
|
|
2W43
 
 | | Structure of L-haloacid dehalogenase from S. tokodaii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HYPOTHETICAL 2-HALOALKANOIC ACID DEHALOGENASE, PHOSPHATE ION | | Authors: | Rye, C.A, Isupov, M.N, Lebedev, A.A, Littlechild, J.A. | | Deposit date: | 2008-11-21 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Biochemical and Structural Studies of a L-Haloacid Dehalogenase from the Thermophilic Archaeon Sulfolobus Tokodaii.
Extremophiles, 13, 2009
|
|
2W11
 
 | | Structure of the L-2-haloacid dehalogenase from Sulfolobus tokodaii | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, 2-HALOALKANOIC ACID DEHALOGENASE | | Authors: | Rye, C.A, Isupov, M.N, Lebedev, A.A, Littlechild, J.A. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Structural Studies of a L-Haloacid Dehalogenase from the Thermophilic Archaeon Sulfolobus Tokodaii.
Extremophiles, 13, 2009
|
|
3HG6
 
 | | Crystal Structure of the Recombinant Onconase from Rana pipiens | | Descriptor: | GLYCEROL, Onconase, SULFATE ION | | Authors: | Camara-Artigas, A, Gavira, J.A, Casares-Atienza, S, Weininger, U, Balbach, J, Garcia-Mira, M.M. | | Deposit date: | 2009-05-13 | | Release date: | 2010-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-state thermal unfolding of onconase.
Biophys.Chem., 159, 2011
|
|
2QI6
 
 | | Crystal structure of protease inhibitor, MIT-2-KB98 in complex with wild type HIV-1 protease | | Descriptor: | N-{(1S,2R)-3-[(1,3-BENZOTHIAZOL-6-YLSULFONYL)(ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYL}-3-HYDROXYBENZAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
3IMW
 
 | | Transthyretin in complex with (E)-2,6-dibromo-4-(2,6-dimethoxystyryl)aniline | | Descriptor: | 2,6-dibromo-4-[(E)-2-(2,6-dimethoxyphenyl)ethenyl]aniline, Transthyretin | | Authors: | Connelly, S, Wilson, I.A. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | A substructure combination strategy to create potent and selective transthyretin kinetic stabilizers that prevent amyloidogenesis and cytotoxicity.
J.Am.Chem.Soc., 132, 2010
|
|
3IMR
 
 | | Transthyretin in complex with (E)-2,6-dibromo-4-(2,6-dichlorostyryl)phenol | | Descriptor: | 2,6-dibromo-4-[(E)-2-(2,6-dichlorophenyl)ethenyl]phenol, Transthyretin | | Authors: | Connelly, S, Wilson, I.A. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A substructure combination strategy to create potent and selective transthyretin kinetic stabilizers that prevent amyloidogenesis and cytotoxicity.
J.Am.Chem.Soc., 132, 2010
|
|
2PB2
 
 | | Structure of biosynthetic N-acetylornithine aminotransferase from Salmonella typhimurium: studies on substrate specificity and inhibitor binding | | Descriptor: | 1,2-ETHANEDIOL, Acetylornithine/succinyldiaminopimelate aminotransferase, GLYCEROL, ... | | Authors: | Rajaram, V, Ratna Prasuna, P, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2007-03-28 | | Release date: | 2007-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of biosynthetic N-acetylornithine aminotransferase from Salmonella typhimurium: Studies on substrate specificity and inhibitor binding
Proteins, 70, 2007
|
|
2QI4
 
 | | Crystal structure of protease inhibitor, MIT-2-AD93 in complex with wild type HIV-1 protease | | Descriptor: | ACETATE ION, N-[(1S,2R)-3-{(1,3-BENZOTHIAZOL-6-YLSULFONYL)[(2S)-2-METHYLBUTYL]AMINO}-1-BENZYL-2-HYDROXYPROPYL]-3-HYDROXYBENZAMIDE, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
3IMS
 
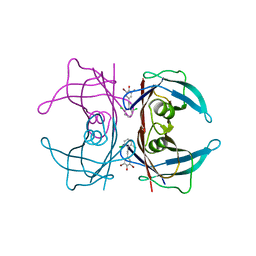 | | Transthyretin in complex with 2,6-dibromo-4-(2,6-dichlorophenethyl)phenol | | Descriptor: | 2,6-dibromo-4-[2-(2,6-dichlorophenyl)ethyl]phenol, Transthyretin | | Authors: | Connelly, S, Wilson, I.A. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A substructure combination strategy to create potent and selective transthyretin kinetic stabilizers that prevent amyloidogenesis and cytotoxicity.
J.Am.Chem.Soc., 132, 2010
|
|
2QI5
 
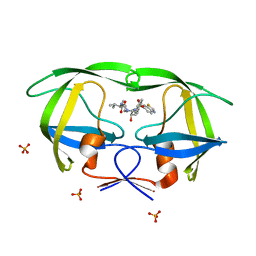 | | Crystal structure of protease inhibitor, MIT-2-KC08 in complex with wild type HIV-1 protease | | Descriptor: | N~2~-ACETYL-N-{(1S,2R)-3-[(1,3-BENZOTHIAZOL-6-YLSULFONYL)(PENTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYL}-L-VALINAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QI3
 
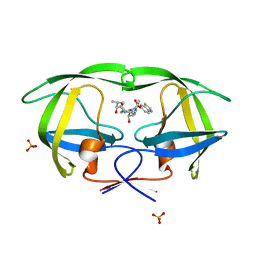 | | Crystal structure of protease inhibitor, MIT-2-AD94 in complex with wild type HIV-1 protease | | Descriptor: | (2S)-N-[(1S,2R)-3-{(1,3-BENZOTHIAZOL-6-YLSULFONYL)[(2S)-2-METHYLBUTYL]AMINO}-1-BENZYL-2-HYDROXYPROPYL]-2-HYDROXY-3-METHYLBUTANAMIDE, PHOSPHATE ION, Protease | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2WGI
 
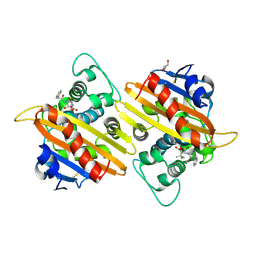 | | Crystal structure of the acyl-enzyme OXA-10 W154A-benzylpenicillin at pH 6 | | Descriptor: | BETA-LACTAMASE OXA-10, GLYCEROL, OPEN FORM - PENICILLIN G | | Authors: | Vercheval, L, Falzone, C, Sauvage, E, Herman, R, Charlier, P, Galleni, M, Kerff, F. | | Deposit date: | 2009-04-20 | | Release date: | 2009-11-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Critical Role of Tryptophan 154 for the Activity and Stability of Class D Beta-Lactamases.
Biochemistry, 48, 2009
|
|
3IMT
 
 | | Transthyretin in complex with (E)-4-(4-aminostyryl)-2,6-dibromophenol | | Descriptor: | 4-[(E)-2-(4-aminophenyl)ethenyl]-2,6-dibromophenol, Transthyretin | | Authors: | Connelly, S, Wilson, I. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A substructure combination strategy to create potent and selective transthyretin kinetic stabilizers that prevent amyloidogenesis and cytotoxicity.
J.Am.Chem.Soc., 132, 2010
|
|
3IIR
 
 | | Crystal Structure of Miraculin like protein from seeds of Murraya koenigii | | Descriptor: | Trypsin inhibitor | | Authors: | Gahloth, D, Selvakumar, P, Shee, C, Kumar, P, Sharma, A.K. | | Deposit date: | 2009-08-03 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cloning, sequence analysis and crystal structure determination of a miraculin-like protein from Murraya koenigii
Arch.Biochem.Biophys., 494, 2010
|
|
2XQB
 
 | | Crystal Structure of anti-IL-15 Antibody in Complex with human IL-15 | | Descriptor: | ANTI-IL-15 ANTIBODY, INTERLEUKIN 15, SULFATE ION | | Authors: | Lowe, D.C, Gerhardt, S, Ward, A, Hargreaves, D, Anderson, M, StGallay, S, Vousden, K, Ferraro, F, Pauptit, R.A, Cochrane, D, Pattison, D.V, Buchanan, C, Popovic, B, Finch, D.K, Wilkinson, T, Sleeman, M, Vaughan, T.J, Cruwys, S, Mallinder, P.R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a High Affinity Anti-Il-15 Antibody: Crystal Structure Reveals an Alpha-Helix in Vh Cdr3 as Key Component of Paratope.
J.Mol.Biol., 406, 2011
|
|
2P6Z
 
 | |
3IMU
 
 | |
2QI1
 
 | | Crystal structure of protease inhibitor, MIT-1-KK81 in complex with wild type HIV-1 protease | | Descriptor: | N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{[(3-METHOXYPHENYL)SULFONYL](2-THIENYLMETHYL)AMINO}PROPYL]-3,4-DIHYDROXYBENZAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2XQR
 
 | | Crystal structure of plant cell wall invertase in complex with a specific protein inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hothorn, M, Van den Ende, W, Lammens, W, Rybin, V, Scheffzek, K. | | Deposit date: | 2010-09-07 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural Insights Into the Ph-Controlled Targeting of Plant Cell-Wall Invertase by a Specific Inhibitor Protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2QHZ
 
 | | Crystal structure of protease inhibitor, MIT-1-AC87 in complex with wild type HIV-1 protease | | Descriptor: | (2E)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{(2-THIENYLMETHYL)[(2,4,5-TRIFLUOROPHENYL)SULFONYL]AMINO}PROPYL]-4,4,4-TRIFLUORO-3-METHYLBUT-2-ENAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QHY
 
 | | Crystal Structure of protease inhibitor, MIT-1-AC86 in complex with wild type HIV-1 protease | | Descriptor: | ACETATE ION, N~2~-ACETYL-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{(2-THIENYLMETHYL)[(2,4,5-TRIFLUOROPHENYL)SULFONYL]AMINO}PROPYL]-L-ALANINAMIDE, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
