6OVO
 
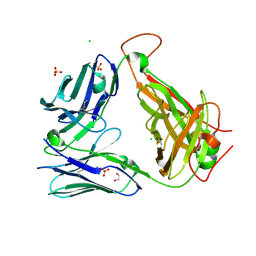 | | Crystal structure of the unliganded PG10 TCR | | Descriptor: | 1,2-ETHANEDIOL, Alpha Chain T-Cell Receptor PG10, Beta Chain T-Cell Receptor PG10, ... | | Authors: | Shahine, A, Rossjohn, J. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | A TCR beta-Chain Motif Biases toward Recognition of Human CD1 Proteins.
J Immunol., 203, 2019
|
|
4X1J
 
 | | X-ray crystal structure of the dimeric BMP antagonist NBL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroblastoma suppressor of tumorigenicity 1 | | Authors: | Thompson, T.B, Nolan, K, Kattamuri, C. | | Deposit date: | 2014-11-24 | | Release date: | 2015-01-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Neuroblastoma Suppressor of Tumorigenicity 1 (NBL1): INSIGHTS FOR THE FUNCTIONAL VARIABILITY ACROSS BONE MORPHOGENETIC PROTEIN (BMP) ANTAGONISTS.
J.Biol.Chem., 290, 2015
|
|
4WKA
 
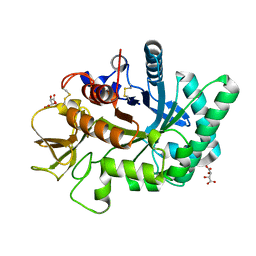 | | Crystal structure of human chitotriosidase-1 catalytic domain at 0.95 A resolution | | Descriptor: | Chitotriosidase-1, L(+)-TARTARIC ACID | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7PA1
 
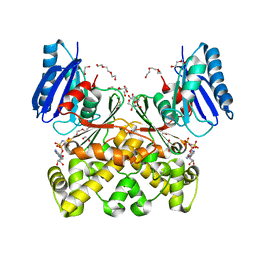 | | Structure of N-acetylglucosamine kinase from Plesiomonas shigelloides in complex with AMP-PNP in the absence of N-acetylglucoseamine substrate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
6OD0
 
 | |
4X1N
 
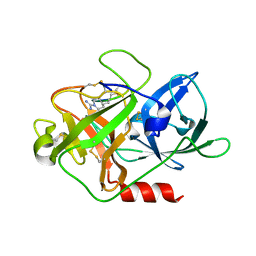 | | The crystal structure of mupain-1-16 in complex with murinised human uPA at pH7.4 | | Descriptor: | Urokinase-type plasminogen activator, mupain-1-16, piperidine-1-carboximidamide | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-25 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
7PUE
 
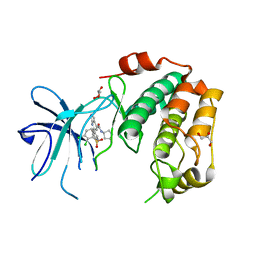 | | Human serum and glucocorticoid-regulated kinase 1 in complex with pyrazolopyridine inhibitor 3a | | Descriptor: | 6-[4-[[2,3-bis(chloranyl)phenyl]sulfonylamino]phenyl]-~{N}-[(3~{R})-pyrrolidin-3-yl]-2~{H}-pyrazolo[3,4-b]pyridine-4-carboxamide, GLYCEROL, Serine/threonine-protein kinase Sgk1 | | Authors: | Dreyer, M.K, Halland, N, Nazare, M. | | Deposit date: | 2021-09-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Rational Design of Highly Potent, Selective, and Bioavailable SGK1 Protein Kinase Inhibitors for the Treatment of Osteoarthritis.
J.Med.Chem., 65, 2022
|
|
377D
 
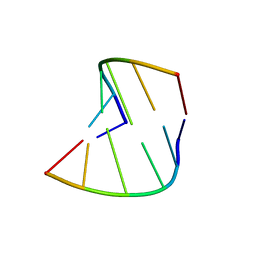 | | 5'-R(*CP*GP*UP*AP*CP*DG)-3' | | Descriptor: | RNA-DNA (5'-R(*CP*GP*UP*AP*CP*DG)-3') | | Authors: | Biswas, R, Mitra, S.N, Sundaralingam, M. | | Deposit date: | 1998-01-26 | | Release date: | 1998-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 1.76 A structure of a pyrimidine start alternating A-RNA hexamer r(CGUAC)dG.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
6OZQ
 
 | |
6PK2
 
 | | CRYSTAL STRUCTURE OF THE CARBOXYLTRANSFERASE SUBUNIT OF ACC (ACCD6) IN COMPLEX WITH INHIBITOR QUIZALOFOP-P derivative FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | 2-{4-[(6-fluoro-1,3-benzothiazol-2-yl)oxy]-2-hydroxyphenyl}-N-methylacetamide, Propionyl-CoA carboxylase subunit beta | | Authors: | Reddy, M.C.M, Nian, Z, Michele, T.C.B, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Elucidating the Inhibition and specificity of binding of
herbicidal aryloxyphenoxypropionates derivatives to Mycobacterium tuberculosis carboxyltransferase
domain of acetyl-coenzyme A(AccD6).
To Be Published
|
|
4WXI
 
 | |
7PWI
 
 | | Structure of the dTDP-sugar epimerase StrM | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, dTDP-4-keto-rhamnose 3,5-epimerase,dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.326 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
4WYO
 
 | |
7Q52
 
 | | Crystal structure of S/T protein kinase PknG from Mycobacterium tuberculosis in complex with inhibitor L2W | | Descriptor: | 2-azanyl-3-(4-fluorophenyl)carbonyl-indolizine-1-carboxamide, FE (III) ION, SODIUM ION, ... | | Authors: | Defelipe, L.A, Burastero, O, Bento, I, Garcia-Alai, M.M. | | Deposit date: | 2021-11-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cosolvent Sites-Based Discovery of Mycobacterium Tuberculosis Protein Kinase G Inhibitors.
J.Med.Chem., 65, 2022
|
|
6OZK
 
 | |
4X1S
 
 | | The crystal structure of mupain-1-16-D9A in complex with murinised human uPA at pH7.4 | | Descriptor: | Urokinase-type plasminogen activator, mupain-1-16, piperidine-1-carboximidamide | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-10-28 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
7PZ4
 
 | | Structure of an LPMO at 2.07x10^4 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | Authors: | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PZ6
 
 | | Structure of an LPMO at 2.22x10^5 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | Authors: | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PZ5
 
 | | Structure of an LPMO at 9.56x10^4 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | Authors: | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PZ8
 
 | | Structure of an LPMO at 3.12x10^6 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | Authors: | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
4X9X
 
 | | Biochemical Roles for Conserved Residues in the Bacterial Fatty Acid Binding Protein Family | | Descriptor: | 1,2-ETHANEDIOL, DegV domain-containing protein MW1315, OLEIC ACID | | Authors: | Broussard, T.C, Miller, D.J, Jackson, P, Nourse, A, Rock, C.O. | | Deposit date: | 2014-12-11 | | Release date: | 2016-01-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Biochemical Roles for Conserved Residues in the Bacterial Fatty Acid-binding Protein Family.
J.Biol.Chem., 291, 2016
|
|
6OCX
 
 | |
6HU6
 
 | | Structure of ARF1 RNA | | Descriptor: | AMMONIUM ION, RNA (19-MER), RNA (5'-R(*CP*UP*GP*GP*UP*AP*CP*UP*C)-3'), ... | | Authors: | Emmerich, C, Lazzaretti, D, Bandholz-Cajamarca, L, Bono, F. | | Deposit date: | 2018-10-05 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | The crystal structure of Staufen1 in complex with a physiological RNA sheds light on substrate selectivity.
Life Sci Alliance, 1, 2018
|
|
6OZN
 
 | |
8GMK
 
 | | Pyruvate bound structure of Citrate Synthase (CitA) in Mycobacterium Tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PYRUVIC ACID, ... | | Authors: | Pathirage, R, Ronning, D, Yamsek, M. | | Deposit date: | 2023-03-26 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Mycobacterium tuberculosis CitA activity is modulated by cysteine oxidation and pyruvate binding.
Rsc Med Chem, 14, 2023
|
|
