8QS0
 
 | |
8QRX
 
 | |
8QRT
 
 | |
8QRH
 
 | | Inactivated tick-borne encephalitis virus (TBEV) vaccine strain Sofjin-Chumakov | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, Small envelope protein M | | Authors: | Moiseenko, A.V, Zhang, Y, Vorovitch, M, Ivanova, A, Liu, Z, Osolodkin, D.I, Egorov, A, Ishmukhametov, A, Sokolova, O.S. | | Deposit date: | 2023-10-07 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The structure of inactivated mature tick-borne encephalitis virus at 3.0 angstrom resolution.
Emerg Microbes Infect, 13, 2024
|
|
8QRG
 
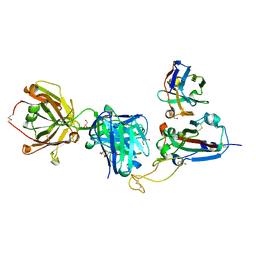 | | SARS-CoV-2 delta RBD complexed with XBB-2 Fab and NbC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NbC1, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QRF
 
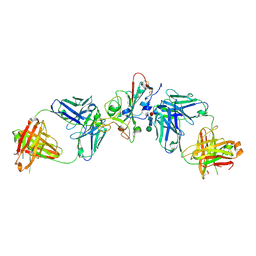 | | SARS-CoV-2 delta RBD complexed with XBB-6 and beta-49 Fabs | | Descriptor: | Beta-49 heavy chain, Beta-49 light chain, Spike protein S1, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QRE
 
 | | Cholera holotoxin (wildtype) | | Descriptor: | ACETATE ION, Cholera enterotoxin subunit A, Cholera enterotoxin subunit B, ... | | Authors: | Mojica, N, Cordara, G, Krengel, U. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Using Vibrio natriegens for High-Yield Production of Challenging Expression Targets and for Protein Perdeuteration.
Biochemistry, 63, 2024
|
|
8QQG
 
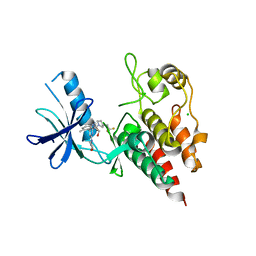 | | Structure of BRAF in Complex With Exarafenib (KIN-2787). | | Descriptor: | (3~{S})-~{N}-[4-methyl-3-[2-morpholin-4-yl-6-[[(2~{R})-1-oxidanylpropan-2-yl]amino]pyridin-4-yl]phenyl]-3-[2,2,2-tris(fluoranyl)ethyl]pyrrolidine-1-carboxamide, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Schmitt, A, Costanzi, E, Kania, R, Chen, Y.K. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.979 Å) | | Cite: | Structure of BRAF in Complex With Exarafenib (KIN-2787).
To Be Published
|
|
8QQF
 
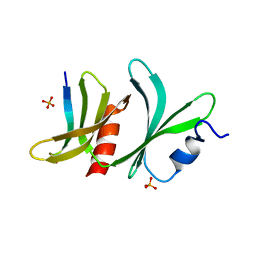 | | TBC1D23 PH domain complexed with STX16 TLY motif | | Descriptor: | GLYCEROL, PHOSPHATE ION, Syntaxin-16, ... | | Authors: | Kaufman, J.G, Owen, D.J. | | Deposit date: | 2023-10-04 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Cargo selective vesicle tethering: The structural basis for binding of specific cargo proteins by the Golgi tether component TBC1D23.
Sci Adv, 10, 2024
|
|
8QQ5
 
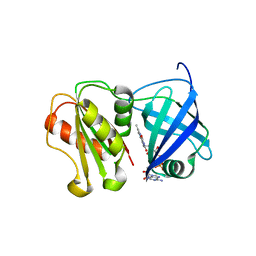 | | Structure of WT SpNox DH domain: a bacterial NADPH oxidase. | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Thepaut, M, Petit-Hartlein, I, Vermot, A, Humm, A.S, Dupeux, F, Marquez, J.A, Smith, S, Fieschi, F. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
8QQ4
 
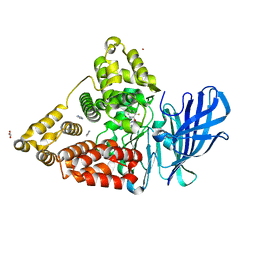 | | LTA4 hydrolase in complex with compound 6(R) | | Descriptor: | (2R)-2-azanyl-3-[5-[4-(5-chloranyl-3-fluoranyl-pyridin-2-yl)oxyphenyl]-1,2,3,4-tetrazol-2-yl]propan-1-ol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2023-10-03 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Amino Alcohols as Highly Potent, Selective, and Orally Efficacious Inhibitors of Leukotriene A4 Hydrolase.
J.Med.Chem., 66, 2023
|
|
8QQ1
 
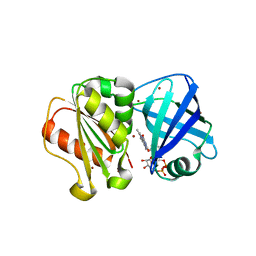 | | SpNOX dehydrogenase domain, mutant F397W in complex with Flavin adenine dinucleotide (FAD) | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Humm, A.S, Dupeux, F, Vermot, A, Petit-Harleim, I, Fieschi, F, Marquez, J.A. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
8QQ0
 
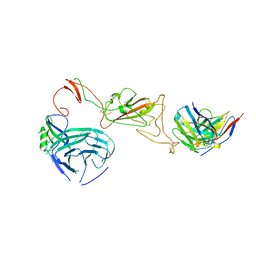 | | SARS-CoV-2 S protein bound to neutralising antibody UZGENT_A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Remaut, H, Reiter, D, Vandenkerckhove, L, Acar, D.D, Witkowski, W, Gerlo, S. | | Deposit date: | 2023-10-03 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Integrating artificial intelligence-based epitope prediction in a SARS-CoV-2 antibody discovery pipeline: caution is warranted.
Ebiomedicine, 100, 2024
|
|
8QPY
 
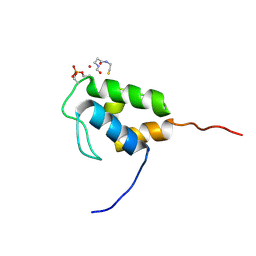 | |
8QPX
 
 | |
8QPR
 
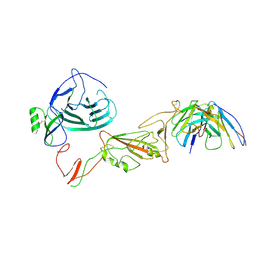 | | SARS-CoV-2 S protein bound to human neutralising antibody UZGENT_G5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG heavy chain - FAB, ... | | Authors: | Remaut, H, Reiter, D, Vandenkerckhove, L, Acar, D.D, Witkowski, W, Gerlo, S. | | Deposit date: | 2023-10-03 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Integrating artificial intelligence-based epitope prediction in a SARS-CoV-2 antibody discovery pipeline: caution is warranted.
Ebiomedicine, 100, 2024
|
|
8QPN
 
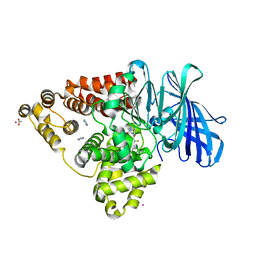 | | LTA4 hydrolase in complex with compound 6(S) | | Descriptor: | (2S)-2-azanyl-3-[5-[4-(5-chloranyl-3-fluoranyl-pyridin-2-yl)oxyphenyl]-1,2,3,4-tetrazol-2-yl]propan-1-ol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2023-10-02 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Amino Alcohols as Highly Potent, Selective, and Orally Efficacious Inhibitors of Leukotriene A4 Hydrolase.
J.Med.Chem., 66, 2023
|
|
8QPK
 
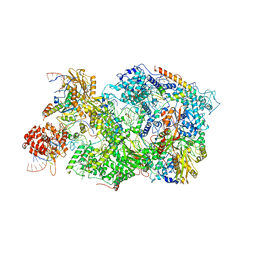 | | Cryo-EM Structure of Pre-B+5'ss Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QPE
 
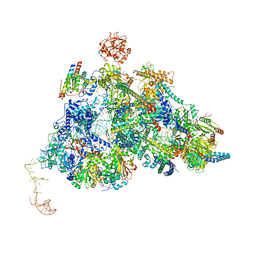 | | Cryo-EM Structure of Pre-B-like Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, INOSITOL HEXAKISPHOSPHATE, Microfibrillar-associated protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-01 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QPC
 
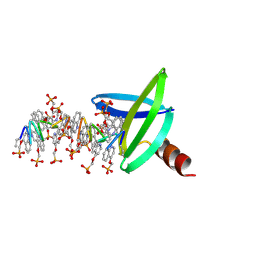 | |
8QPB
 
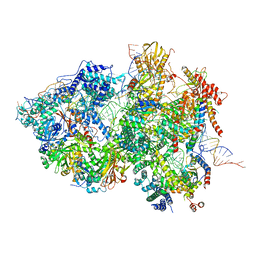 | | Cryo-EM Structure of Pre-B+ATP Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-01 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QPA
 
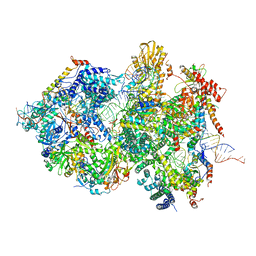 | | Cryo-EM Structure of Pre-B+5'ssLNG Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-01 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QP9
 
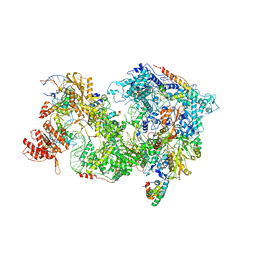 | | Cryo-EM Structure of Pre-B+AMPPNP Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, INOSITOL HEXAKISPHOSPHATE, Pre-mRNA-processing factor 6, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-09-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QP8
 
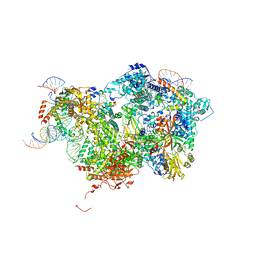 | | Cryo-EM Structure of Pre-B Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, INOSITOL HEXAKISPHOSPHATE, Pre-mRNA-processing factor 6, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-09-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QOZ
 
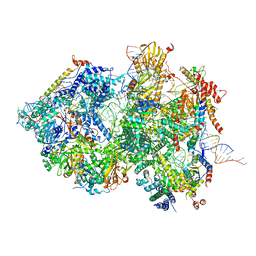 | | Cryo-EM Structure of Pre-B+5'ss+ATPgammaS Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss RNA oligo, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-09-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
