4GD7
 
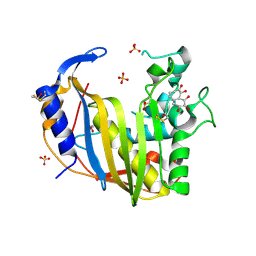 | | Wild-Type Human Thymidylate Synthase with bound Purpurogallin | | Descriptor: | 1,2-ETHANEDIOL, 2,3,4,6-tetrahydroxy-5H-benzo[7]annulen-5-one, SULFATE ION, ... | | Authors: | Celeste, L.R, Lebioda, L. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Oxidation of Cysteine 195 of Huyman Thymidylate Synthase by Purpurogallin
To be published, 2012
|
|
4FGT
 
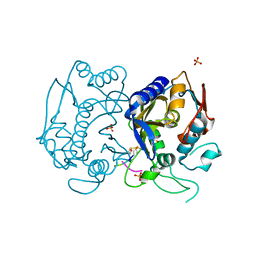 | | Allosteric peptidic inhibitor of human thymidylate synthase that stabilizes inactive conformation of the enzyme. | | Descriptor: | CG peptide, SULFATE ION, Thymidylate synthase | | Authors: | Tochowicz, A, Finer-Moore, J, Stroud, R.M, Costi, M.P. | | Deposit date: | 2012-06-04 | | Release date: | 2013-03-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Alanine mutants of the interface residues of human thymidylate synthase decode key features of the binding mode of allosteric anticancer peptides.
J.Med.Chem., 58, 2015
|
|
3HPW
 
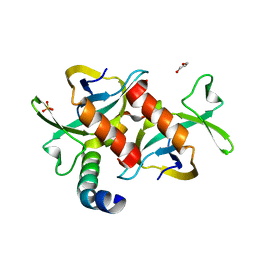 | | CcdB dimer in complex with one C-terminal CcdA domain | | Descriptor: | Cytotoxic protein ccdB, DI(HYDROXYETHYL)ETHER, Protein ccdA, ... | | Authors: | De Jonge, N, Loris, R, Garcia-Pino, A, Buts, L. | | Deposit date: | 2009-06-05 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | Rejuvenation of CcdB-Poisoned Gyrase by an Intrinsically Disordered Protein Domain.
Mol.Cell, 35, 2009
|
|
4JEF
 
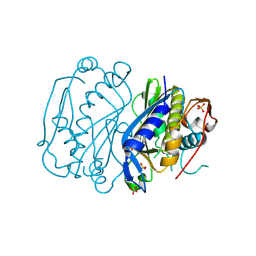 | |
4IEJ
 
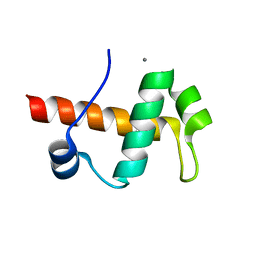 | |
4GYH
 
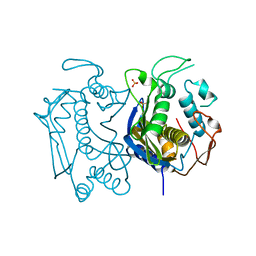 | |
4KPW
 
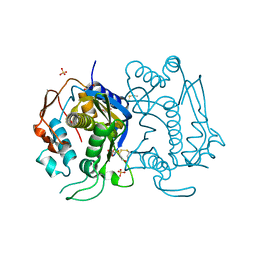 | |
4I80
 
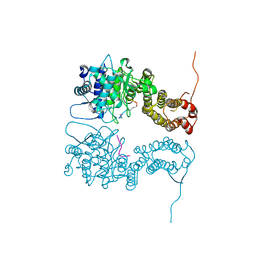 | |
4H1I
 
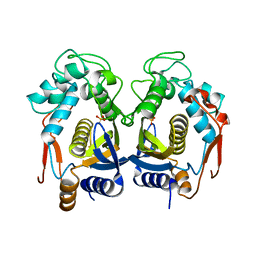 | |
1ZVV
 
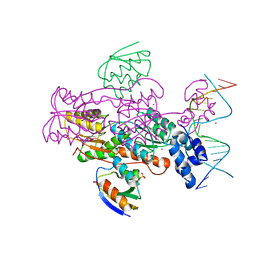 | | Crystal structure of a ccpa-crh-dna complex | | Descriptor: | DNA recognition strand CRE, Glucose-resistance amylase regulator, HPr-like protein crh, ... | | Authors: | Schumacher, M.A, Brennan, R.G, Hillen, W, Seidel, G. | | Deposit date: | 2005-06-02 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Phosphoprotein Crh-Ser46-P displays altered binding to CcpA to effect carbon catabolite regulation.
J.Biol.Chem., 281, 2006
|
|
8J6W
 
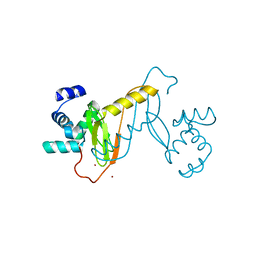 | |
3U88
 
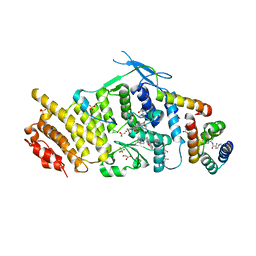 | | Crystal structure of human menin in complex with MLL1 and LEDGF | | Descriptor: | (4beta,8alpha,9R)-6'-methoxy-10,11-dihydrocinchonan-9-ol, CHOLIC ACID, GLYOXYLIC ACID, ... | | Authors: | Huang, J, Wan, B, Lei, M. | | Deposit date: | 2011-10-16 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The same pocket in menin binds both MLL and JUND but has opposite effects on transcription.
Nature, 482, 2012
|
|
6ZXO
 
 | |
4WQO
 
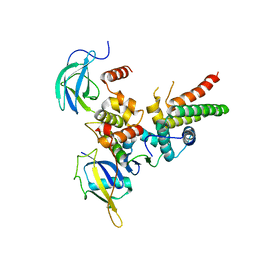 | | Structure of VHL-EloB-EloC-Cul2 | | Descriptor: | Cullin-2, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Nguyen, H.C, Xiong, Y. | | Deposit date: | 2014-10-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Insights into Cullin-RING E3 Ubiquitin Ligase Recruitment: Structure of the VHL-EloBC-Cul2 Complex.
Structure, 23, 2015
|
|
2FO1
 
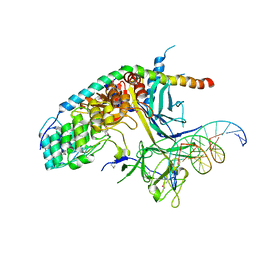 | | Crystal Structure of the CSL-Notch-Mastermind ternary complex bound to DNA | | Descriptor: | 5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3', 5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3', Lin-12 and glp-1 phenotype protein 1, ... | | Authors: | Wilson, J.J, Kovall, R.A. | | Deposit date: | 2006-01-12 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA.
Cell(Cambridge,Mass.), 124, 2006
|
|
8OJ3
 
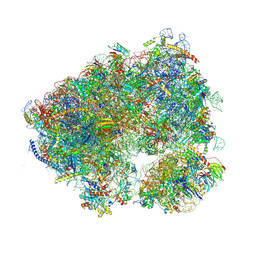 | | Crystal structure of the Candida albicans 80S ribosome in complex with geneticin G418 (rotated state) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-23 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Drug-induced rotational movement of the ribosome is a key factor for read-through enhancement
To Be Published
|
|
8OEQ
 
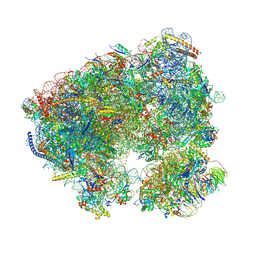 | | Crystal structure of the Candida albicans 80S ribosome in complex with Paromomycin (250uM) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-12 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Drug-induced rotational movement of the ribosome is a key factor for read-through enhancement
To Be Published
|
|
2VF0
 
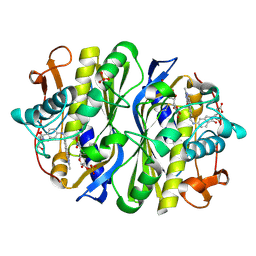 | | CRYSTAL STRUCTURE OF THE THYMIDYLATE SYNTHASE K48Q COMPLEXED WITH 5NO2DUMP AND BW1843U89 | | Descriptor: | 2'-DEOXY-5-NITROURIDINE 5'-MONOPHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, SULFATE ION, ... | | Authors: | Sotelo-Mundo, R.R, Arreola, R, Maley, F, Montfort, W.R. | | Deposit date: | 2007-10-27 | | Release date: | 2007-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Role of an Invariant Lysine Residue in Folate Binding on Escherichia Coli Thymidylate Synthase: Calorimetric and Crystallographic Analysis of the K48Q Mutant.
Int.J.Biochem.Cell Biol., 40, 2008
|
|
2VET
 
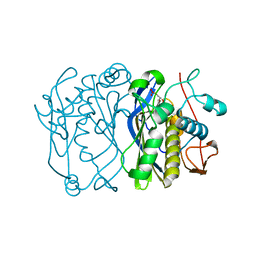 | | CRYSTAL STRUCTURE OF THE THYMIDYLATE SYNTHASE K48Q COMPLEXED WITH DUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Sotelo-Mundo, R.R, Arreola, R, Maley, F, Montfort, W.R. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of an Invariant Lysine Residue in Folate Binding on Escherichia Coli Thymidylate Synthase: Calorimetric and Crystallographic Analysis of the K48Q Mutant.
Int.J.Biochem.Cell Biol., 40, 2008
|
|
3U85
 
 | |
6OJV
 
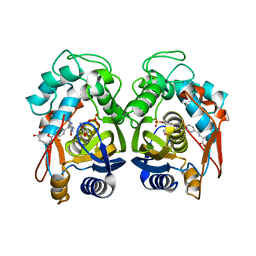 | | Crystal structure of human thymidylate synthase delta(7-29) in complex with dUMP and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-L-glutamic acid | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, N-{4-[(2-amino-4-hydroxy-7H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzoyl}-L-glutamic acid, Thymidylate synthase,Thymidylate synthase | | Authors: | Czyzyk, D.J, Anderson, K.S, Valhondo, M, Jorgensen, W.L. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Understanding the structural basis of species selective, stereospecific inhibition for Cryptosporidium and human thymidylate synthase.
Febs Lett., 593, 2019
|
|
6OJU
 
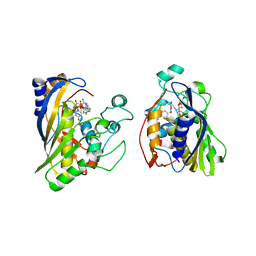 | | Crystal structure of human thymidylate synthase Delta (7-29) in complex with dUMP and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-D-glutamic acid | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, N-{4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}-D-glutamic acid, Thymidylate synthase,Thymidylate synthase | | Authors: | Czyzyk, D.J, Anderson, K.S, Valhondo, M, Jorgensen, W.L. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Understanding the structural basis of species selective, stereospecific inhibition for Cryptosporidium and human thymidylate synthase.
Febs Lett., 593, 2019
|
|
3C0Y
 
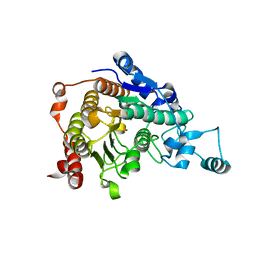 | | Crystal structure of catalytic domain of human histone deacetylase HDAC7 | | Descriptor: | Histone deacetylase 7a, POTASSIUM ION, ZINC ION | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Loppnau, P, Kwiatkowski, N.P, Mazitschek, R, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity.
J.Biol.Chem., 283, 2008
|
|
3F9W
 
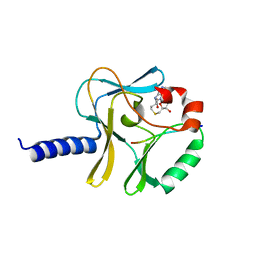 | | Structural Insights into Lysine Multiple Methylation by SET Domain Methyltransferases, SET8-Y334F / H4-Lys20 / AdoHcy | | Descriptor: | Histone H4, Histone-lysine N-methyltransferase SETD8, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Couture, J.-F, Dirk, L.M.A, Brunzelle, J.S, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural origins for the product specificity of SET domain protein methyltransferases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3F9Y
 
 | | Structural Insights into Lysine Multiple Methylation by SET Domain Methyltransferases, SET8-Y334F / H4-Lys20me1 / AdoHcy | | Descriptor: | Histone H4, Histone-lysine N-methyltransferase SETD8, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Couture, J.-F, Dirk, L.M.A, Brunzelle, J.S, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural origins for the product specificity of SET domain protein methyltransferases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
