7YMD
 
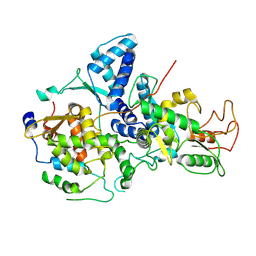 | | Cryo-EM structure of Nse1/3/4 | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, Non-structural maintenance of chromosomes element 1 | | Authors: | Qian, L, Jun, Z, Zhenguo, C, Wang, L. | | Deposit date: | 2022-07-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.176 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
7FJ1
 
 | | Cryo-EM structure of pseudorabies virus C-capsid | | Descriptor: | Capsid vertex component 1, DNA packaging tegument protein UL25, Major capsid protein, ... | | Authors: | Zheng, Q, Li, S, Zha, Z, Sun, H. | | Deposit date: | 2021-08-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structures of pseudorabies virus capsids.
Nat Commun, 13, 2022
|
|
1XPR
 
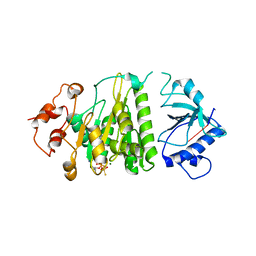 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic 5a-formylbicyclomycin (FB) | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', 5A-FORMYLBICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
2B5M
 
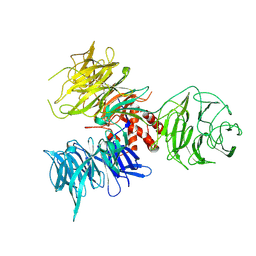 | | Crystal Structure of DDB1 | | Descriptor: | damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
1UE2
 
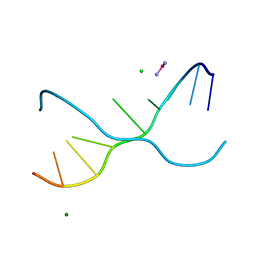 | | Crystal structure of d(GC38GAAAGCT) | | Descriptor: | 5'-D(*GP*(C38)P*GP*AP*AP*AP*GP*CP*T)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanaba, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-05-08 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of d(GCGAAAGC) (hexagonal form): a base-intercalated duplex as a stable structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UB8
 
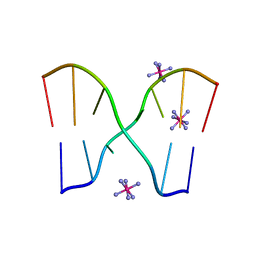 | | Crystal structure of d(GCGAAGC), bending duplex with a bulge-in residue | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*GP*C)-3', COBALT HEXAMMINE(III) | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-03-31 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of d(GCGAAGC) and d(GCGAAAGC) (tetragonal form): a switching of partners of the sheared G.A pairs to form a functional G.AxA.G crossing.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1F3U
 
 | |
4MQ3
 
 | |
4LEU
 
 | | Crystal Structure of THA8-like protein from Arabidopsis thaliana | | Descriptor: | Pentatricopeptide repeat-containing protein At3g46870 | | Authors: | Ke, J, Chen, R.Z, Ban, T, Brunzelle, J.S, Gu, X, Kang, Y, Melcher, K, Zhu, J.K, Xu, H.E. | | Deposit date: | 2013-06-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a PLS-class Pentatricopeptide Repeat Protein Provides Insights into Mechanism of RNA Recognition.
J.Biol.Chem., 288, 2013
|
|
1UE4
 
 | | Crystal structure of d(GCGAAAGC) | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*AP*GP*C)-3', MAGNESIUM ION | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-05-09 | | Release date: | 2004-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of d(GCGAAGC) and d(GCGAAAGC) (tetragonal form): a switching of partners of the sheared G.A pairs to form a functional G.AxA.G crossing.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1U01
 
 | | High resolution NMR structure of 5-d(GCGT*GCG)-3/5-d(CGCACGC)-3 (T*represents a cyclohexenyl nucleotide) | | Descriptor: | 5'-D(*CP*GP*CP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*(XTR)P*GP*CP*G)-3' | | Authors: | Nauwelaerts, K, Lescrinier, E, Sclep, G, Herdewijn, P. | | Deposit date: | 2004-07-12 | | Release date: | 2005-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Cyclohexenyl nucleic acids: conformationally flexible oligonucleotides.
Nucleic Acids Res., 33, 2005
|
|
1UE3
 
 | | Crystal structure of d(GCGAAAGC) containing hexaamminecobalt | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*AP*GP*C)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-05-08 | | Release date: | 2004-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of d(GCGAAAGC) (hexagonal form): a base-intercalated duplex as a stable structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4ID1
 
 | |
1HAR
 
 | |
5W45
 
 | | Crystal structure of APOBEC3H | | Descriptor: | APOBEC3H, ZINC ION | | Authors: | Ito, F, Yang, H.J, Xiao, X, Li, S.X, Wolfe, A, Zirkle, B, Arutiunian, V, Chen, X.S. | | Deposit date: | 2017-06-09 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Understanding the Structure, Multimerization, Subcellular Localization and mC Selectivity of a Genomic Mutator and Anti-HIV Factor APOBEC3H.
Sci Rep, 8, 2018
|
|
1E2F
 
 | | Human thymidylate kinase complexed with thymidine monophosphate, adenosine diphosphate and a magnesium-ion | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Ostermann, N, Schlichting, I, Brundiers, R, Konrad, M, Reinstein, J, Veit, T, Goody, R.S, Lavie, A. | | Deposit date: | 2000-05-22 | | Release date: | 2001-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights Into the Phosphoryltransfer Mechanism of Human Thymidylate Kinase Gained from Crystal Structures of Enzyme Complexes Along the Reaction Coordinate
Structure, 8, 2000
|
|
4NFT
 
 | | Crystal structure of human lnkH2B-h2A.Z-Anp32e | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member E, Histone H2B type 2-E, Histone H2A.Z | | Authors: | Shan, S, Pan, L, Mao, Z, Wang, W, Sun, J, Dong, Q, Liang, X, Ding, X, Chen, S, Dai, L, Zhang, Z, Zhu, B, Zhou, Z. | | Deposit date: | 2013-11-01 | | Release date: | 2014-04-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z
Cell Res., 24, 2014
|
|
4N0L
 
 | | Methanopyrus kandleri Csm3 crystal structure | | Descriptor: | Predicted component of a thermophile-specific DNA repair system, contains a RAMP domain, ZINC ION | | Authors: | Hrle, A, Su, A.A, Ebert, J, Benda, C, Conti, E, Randau, L. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure and RNA-binding properties of the Type III-A CRISPR-associated protein Csm3.
Rna Biol., 10, 2013
|
|
4HEJ
 
 | | Discovery of Selective and Potent Inhibitors of Gram-positive Bacterial Thymidylate Kinase (TMK): Compund 16 | | Descriptor: | 5-methyl-1-[(3S)-1-{3-[3-(trifluoromethyl)phenoxy]benzyl}piperidin-3-yl]pyrimidine-2,4(1H,3H)-dione, Thymidylate kinase | | Authors: | Martinez-Botella, G, Breen, J, Duffy, J, Dumas, J, Geng, B, Gowers, I, Green, O, Guler, S, Hentemann, M, Hernandez-Juan, F, Joseph-McCarthy, D, Kawatkar, S, Larsen, N, Lazari, O, Loch, J, Macritchie, J. | | Deposit date: | 2012-10-03 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Selective and Potent Inhibitors of Gram-Positive Bacterial Thymidylate Kinase (TMK).
J.Med.Chem., 55, 2012
|
|
4NJC
 
 | |
1Y98
 
 | | Structure of the BRCT repeats of BRCA1 bound to a CtIP phosphopeptide. | | Descriptor: | Breast cancer type 1 susceptibility protein, COBALT (II) ION, CtIP PHOSPHORYLATED PEPTIDE, ... | | Authors: | Varma, A.K, Brown, R.S, Birrane, G, Ladias, J.A.A. | | Deposit date: | 2004-12-14 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Cell Cycle Checkpoint Control by the BRCA1-CtIP Complex.
Biochemistry, 44, 2005
|
|
4N7R
 
 | | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its binding protein | | Descriptor: | Genomic DNA, chromosome 3, P1 clone: MXL8, ... | | Authors: | Zhao, A, Fang, Y, Lin, Y, Gong, W, Liu, L. | | Deposit date: | 2013-10-16 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its stimulator protein
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5YY9
 
 | | Crystal structure of Tandem Tudor Domain of human UHRF1 in complex with LIG1-K126me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Ligase 1 | | Authors: | Kori, S, Defossez, P.A, Arita, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
5YYA
 
 | | Crystal structure of Tandem Tudor Domain of human UHRF1 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, SULFATE ION | | Authors: | Kori, S, Defossez, P.A, Arita, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
4I1A
 
 | | Crystal Structure of the Apo Form of RapI | | Descriptor: | CHLORIDE ION, Response regulator aspartate phosphatase I | | Authors: | Parashar, V, Jeffrey, P.D, Neiditch, M.B. | | Deposit date: | 2012-11-20 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.443 Å) | | Cite: | Conformational change-induced repeat domain expansion regulates rap phosphatase quorum-sensing signal receptors.
Plos Biol., 11, 2013
|
|
