6DOX
 
 | |
6DPB
 
 | |
1KLC
 
 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1KLD
 
 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 18-33 OF 33 STRUCTURES | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1UB3
 
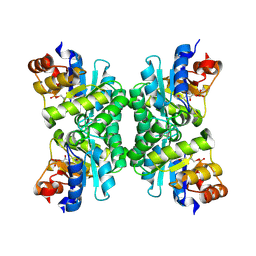 | | Crystal Structure of Tetrameric Structure of Aldolase from thermus thermophilus HB8 | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, Aldolase protein | | Authors: | Lokanath, N.K, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-28 | | Release date: | 2003-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6DKJ
 
 | | human GIPR ECD and Fab complex | | Descriptor: | 1,2-ETHANEDIOL, Fab heavy chain, Fab light chain, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Anti-obesity effects of GIPR antagonists alone and in combination with GLP-1R agonists in preclinical models.
Sci Transl Med, 10, 2018
|
|
2Y85
 
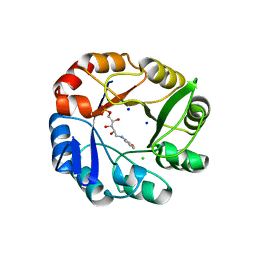 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS PHOSPHORIBOSYL ISOMERASE WITH BOUND RCDRP | | Descriptor: | 1-(O-CARBOXY-PHENYLAMINO)-1-DEOXY-D-RIBULOSE-5-PHOSPHATE, CHLORIDE ION, PHOSPHORIBOSYL ISOMERASE A, ... | | Authors: | Kuper, J, Geerlof, A, Wilmanns, M. | | Deposit date: | 2011-02-03 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bisubstrate Specificity in Histidine/Tryptophan Biosynthesis Isomerase from Mycobacterium Tuberculosis by Active Site Metamorphosis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6DOV
 
 | |
6DP8
 
 | |
6DPP
 
 | |
8QV6
 
 | | Structure of human SPNS2 in DDM | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody D12, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Li, H.Z, Pike, A.C.W, McKinley, G, Mukhopadhyay, S.M.M, Moreau, C, Scacioc, A, Abrusci, P, Borkowska, O, Chalk, R, Stefanic, S, Burgess-Brown, N, Duerr, K.L, Sauer, D.B. | | Deposit date: | 2023-10-17 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Transport and inhibition of the sphingosine-1-phosphate exporter SPNS2.
Nat Commun, 16, 2025
|
|
8QV5
 
 | | Structure of human SPNS2 in LMNG | | Descriptor: | Nanobody D12, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Li, H.Z, Pike, A.C.W, McKinley, G, Mukhopadhyay, S.M.M, Moreau, C, Scacioc, A, Abrusci, P, Borkowska, O, Chalk, R, Stefanic, S, Burgess-Brown, N, Duerr, K.L, Sauer, D.B. | | Deposit date: | 2023-10-17 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Transport and inhibition of the sphingosine-1-phosphate exporter SPNS2.
Nat Commun, 16, 2025
|
|
6DOF
 
 | |
6DOT
 
 | |
9CSE
 
 | | Crystal structure of Repeats-in-Toxin-like domain from Aeromonas hydrophila | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Large adhesion protein | | Authors: | Ye, Q, Vance, T.D.R, Davies, P.L. | | Deposit date: | 2024-07-23 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Aeromonas hydrophila RTX adhesin has three ligand-binding domains that give the bacterium the potential to adhere to and aggregate a wide variety of cell types.
Mbio, 16, 2025
|
|
6DPA
 
 | |
6DPL
 
 | |
2YLF
 
 | |
2YFN
 
 | | galactosidase domain of alpha-galactosidase-sucrose kinase, AgaSK | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-GALACTOSIDASE-SUCROSE KINASE AGASK, GLYCEROL, ... | | Authors: | Sulzenbacher, G, Bruel, L, Tison-Cervera, M, Pujol, A, Nicoletti, C, Perrier, J, Galinier, A, Ropartz, D, Fons, M, Pompeo, F, Giardina, T. | | Deposit date: | 2011-04-07 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Agask, a Bifunctional Enzyme from the Human Microbiome Coupling Galactosidase and Kinase Activities
J.Biol.Chem., 286, 2011
|
|
4MS7
 
 | | Crystal structure of Schizosaccharomyces pombe sst2 catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AMSH-like protease sst2, ... | | Authors: | Shrestha, R.K, Ronau, J.A, Das, C. | | Deposit date: | 2013-09-18 | | Release date: | 2014-06-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Insights into the Mechanism of Deubiquitination by JAMM Deubiquitinases from Cocrystal Structures of the Enzyme with the Substrate and Product.
Biochemistry, 53, 2014
|
|
2YKK
 
 | | Structure of a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CEL44C, ... | | Authors: | Ariza, A, Eklof, J.M, Spadiut, O, Offen, W.A, Roberts, S.M, Besenmatter, W, Friis, E.P, Skjot, M, Wilson, K.S, Brumer, H, Davies, G. | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and Activity of Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44.
J.Biol.Chem., 286, 2011
|
|
3HFD
 
 | |
7OOK
 
 | | Bacteriophage PRD1 Major Capsid Protein P3 in complex with CPZ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, CHLORIDE ION, ... | | Authors: | Duyvesteyn, H.M.E, Peccati, F, Martinez-Castillo, A, Jimenez-Oses, G, Oksanen, H.M, Stuart, D.I, Abrescia, N.G.A. | | Deposit date: | 2021-05-27 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Bacteriophage PRD1 as a nanoscaffold for drug loading
Nanoscale, 13, 2021
|
|
2NPW
 
 | | Solution Structures of a DNA Dodecamer Duplex with a Cisplatin 1,2-d(GG) Intrastrand Cross-Link | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', Cisplatin | | Authors: | Wu, Y, Bhattacharyya, D, Chaney, S, Campbell, S. | | Deposit date: | 2006-10-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a DNA Dodecamer Duplex with and without a Cisplatin 1,2-d(GG) Intrastrand Cross-Link: Comparison with the Same DNA Duplex Containing an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link
Biochemistry, 46, 2007
|
|
6E1M
 
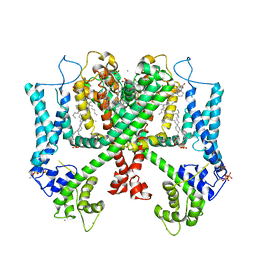 | | Structure of AtTPC1(DDE) reconstituted in saposin A | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CALCIUM ION, PALMITIC ACID, ... | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
