6L4N
 
 | |
6L4I
 
 | |
5FI3
 
 | | HETEROYOHIMBINE SYNTHASE THAS1 FROM CATHARANTHUS ROSEUS - COMPLEX WITH NADP+ | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Tetrahydroalstonine synthase, ... | | Authors: | Stavrinides, A, Tatsis, E.C, Caputi, L, Foureau, E, Stevenson, C.E.M, Lawson, D.M, Courdavault, V, O'Connor, S.E. | | Deposit date: | 2015-12-22 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity.
Nat Commun, 7, 2016
|
|
5FJ2
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 7-((4-CHLORO-3-((METHYLAMINO)METHYL) PHENOXY)METHYL)QUINOLIN-2-AMINE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[4-chloranyl-3-(methylaminomethyl)phenoxy]methyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-10-06 | | Release date: | 2015-10-28 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Phenyl Ether- and Aniline-Containing 2-Aminoquinolines as Potent and Selective Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
5FJI
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5FK7
 
 | |
5FF8
 
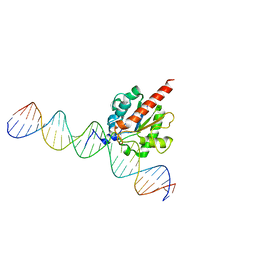 | | TDG enzyme-product complex | | Descriptor: | DNA, G/T mismatch-specific thymine DNA glycosylase | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
5FFJ
 
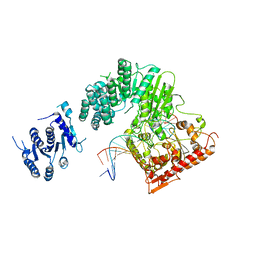 | |
5V92
 
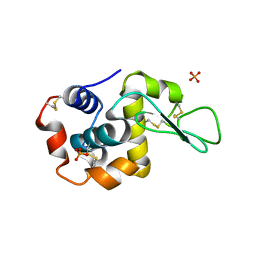 | |
5FGS
 
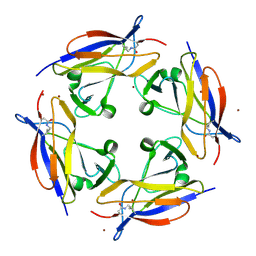 | | Crystal structure of C-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG - P21212 space group | | Descriptor: | Cell surface protein SpaA, ZINC ION | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-21 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5UYR
 
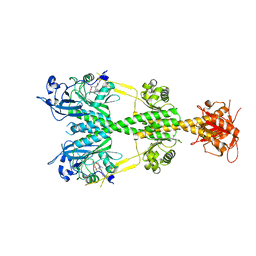 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP mutant D199A from Xanthomonas campestris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Klinke, S, Goldbaum, F.A, Bonomi, H.R. | | Deposit date: | 2017-02-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Pr-favoured variants of the bacteriophytochrome from the plant pathogen Xanthomonas campestris hint on light regulation of virulence-associated mechanisms.
Febs J., 288, 2021
|
|
5FGZ
 
 | | E. coli PBP1b in complex with FPI-1465 | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 1B, [[(3~{R},6~{S})-1-methanoyl-6-[[(3~{S})-pyrrolidin-3-yl]oxycarbamoyl]piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-12-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5VA6
 
 | | CRYSTAL STRUCTURE OF ATXR5 IN COMPLEX WITH HISTONE H3.1 MONO-METHYLATED ON R26 | | Descriptor: | Histone H3.1, Probable Histone-lysine N-methyltransferase ATXR5, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
6LL9
 
 | |
5FR2
 
 | | Farnesylated RhoA-GDP in complex with RhoGDI-alpha, lysine acetylated at K178 | | Descriptor: | FARNESYL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kuhlmann, N, Wroblowski, S, Knyphausen, P, de Boor, S, Brenig, J, Zienert, A.Y, Meyer-Teschendorf, K, Praefcke, G.J.K, Nolte, H, Krueger, M, Schacherl, M, Baumann, U, James, L.C, Chin, J.W, Lammers, M. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural and Mechanistic Insights Into the Regulation of the Fundamental Rho-Regulator Rhogdi Alpha by Lysine Acetylation.
J.Biol.Chem., 291, 2016
|
|
5FRE
 
 | | Characterization of a novel CBM from Clostridium perfringens | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Ribeiro, J, Pau, W, Pifferi, C, Renaudet, O, Varrot, A, Mahal, L.K, Imberty, A. | | Deposit date: | 2015-12-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a High-Affinity Sialic Acid-Specific Cbm40 from Clostridium Perfringens and Engineering of a Divalent Form.
Biochem.J., 473, 2016
|
|
5FHH
 
 | | Structure of human Pif1 helicase domain residues 200-641 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, TETRAFLUOROALUMINATE ION | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
6LOP
 
 | | Crystal Structure of Class IB terpene synthase bound with geranylgeraniol | | Descriptor: | (2~{E},6~{E},10~{E})-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-ol, Tetraprenyl-beta-curcumene synthase | | Authors: | Fujihashi, M, Inagi, H, Miki, K. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Characterization of Class IB Terpene Synthase: The First Crystal Structure Bound with a Substrate Surrogate.
Acs Chem.Biol., 15, 2020
|
|
6LOX
 
 | | Crystal Structure of human glutaminase with macrocyclic inhibitor | | Descriptor: | (E)-15,22-Dioxa-4,11-diaza-5(2,5)-thiadiazola-10(3,6)-pyridazina-1,14(1,3)-dibenzenacyclodocosaphan-18-ene-3,12-dione, Glutaminase kidney isoform, mitochondrial | | Authors: | Bian, J, Li, Z, Xu, X, Wang, J, Li, L. | | Deposit date: | 2020-01-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Enabled Discovery of Novel Macrocyclic Inhibitors Targeting Glutaminase 1 Allosteric Binding Site.
J.Med.Chem., 64, 2021
|
|
5FNS
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3s)-{4-Chloro-3-[(N-methylmethanesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl) propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FWI
 
 | |
6LKC
 
 | | Crystal structure of PfaD from Shewanella piezotolerans in complex with FMN | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Zhang, M.L, Li, Q, Meng, S.S, Guo, L.J, He, L, Huang, J.Z, Li, L, Zhang, H.D. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural Insights into the Trans -Acting Enoyl Reductase in the Biosynthesis of Long-Chain Polyunsaturated Fatty Acids in Shewanella piezotolerans .
J.Agric.Food Chem., 69, 2021
|
|
5FVB
 
 | | CRYSTAL STRUCTURE OF PHORMIDIUM C-PHYCOERYTHRIN AT PH 5.0 | | Descriptor: | C-PHYCOERYTHRIN ALPHA SUBUNIT, C-PHYCOERYTHRIN BETA SUBUNIT, GLYCEROL, ... | | Authors: | Kumar, V, Sonani, R.R, Sharma, M, Gupta, G.D, Madamwar, D. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure Analysis of C-Phycoerythrin from Marine Cyanobacterium Phormidium Sp. A09Dm.
Photosynth.Res., 129, 2016
|
|
5FX5
 
 | | Novel inhibitors of human rhinovirus 3C protease | | Descriptor: | GLYCEROL, RHINOVIRUS 3C PROTEASE, TETRAETHYLENE GLYCOL, ... | | Authors: | Kawatkar, S, Ek, M, Hoesch, V, Gagnon, M, Nilsson, E, Lister, T, Olsson, L, Patel, J, Yu, Q. | | Deposit date: | 2016-02-24 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Structure-Activity Relationships of Novel Inhibitors of Human Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5UNS
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-[(3-Ethyl-5-((methylamino)methyl)phenoxy)methyl]quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-({3-ethyl-5-[(methylamino)methyl]phenoxy}methyl)quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
