2BSL
 
 | | Crystal structure of L. lactis dihydroorotate dehydrogense A in complex with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE A, ... | | Authors: | Wolfe, A.E, Hansen, M, Gattis, S.G, Hu, Y.-C, Johansson, E, Arent, S, Larsen, S, Palfey, B.A. | | Deposit date: | 2005-05-23 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interaction of Benzoate Pyrimidine Analogues with Class 1A Dihydroorotate Dehydrogenase from Lactococcus Lactis.
Biochemistry, 46, 2007
|
|
5AOY
 
 | | Structure of mouse Endonuclease V | | Descriptor: | ENDONUCLEASE V | | Authors: | Vik, E.S, Nawaz, M.S, Ronander, M.E, Solvoll, A.M, Strom-Andersen, P, Bjoras, M, Alseth, I, Dalhus, B. | | Deposit date: | 2015-09-14 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Md Simulation of Mouse Endov Reveal Wedge Motif Plasticity in This Inosine-Specific Endonuclease.
Sci.Rep., 6, 2016
|
|
2BP1
 
 | | Structure of the aflatoxin aldehyde reductase in complex with NADPH | | Descriptor: | AFLATOXIN B1 ALDEHYDE REDUCTASE MEMBER 2, CITRATE ANION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Debreczeni, J.E, Lukacik, P, Kavanagh, K, Dubinina, E, Bray, J, Colebrook, S, Haroniti, A, Edwards, A, Arrowsmith, C, Sundstrom, M, von Delft, F, Gileadi, O, Oppermann, U. | | Deposit date: | 2005-04-17 | | Release date: | 2005-05-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Aflatoxin Aldehyde Reductase in Complex with Nadph
To be Published
|
|
8CU6
 
 | | Crystal structure of A2AAR-StaR2-S277-bRIL in complex with a novel A2a antagonist, LJ-4517 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3R,4R)-2-[(8P)-6-amino-2-(hex-1-yn-1-yl)-8-(thiophen-2-yl)-9H-purin-9-yl]oxolane-3,4-diol, Adenosine receptor A2a,Soluble cytochrome b562, ... | | Authors: | Shiriaeva, A, Park, D.-J, Kim, G, Lee, Y, Hou, X, Jarhad, D.B, Kim, G, Yu, J, Hyun, Y.E, Kim, W, Gao, Z.-G, Jacobson, K.A, Han, G.W, Stevens, R.C, Jeong, L.S, Choi, S, Cherezov, V. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GPCR Agonist-to-Antagonist Conversion: Enabling the Design of Nucleoside Functional Switches for the A 2A Adenosine Receptor.
J.Med.Chem., 65, 2022
|
|
5NCE
 
 | | Structure of PsDef1 defensin from Pinus sylvestris | | Descriptor: | Defensin-1 | | Authors: | Khairutdinov, B.I, Ermakova, E.A, Bessolitsyna, E.K, Toporkova, Y.Y, Tarasova, N.B, Kovaleva, V, Zuev, Y.F, Nesmelova, I.V. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure, conformational dynamics, and biological activity of PsDef1 defensin from Pinus sylvestris.
Biochim. Biophys. Acta, 1865, 2017
|
|
6E26
 
 | |
2IF7
 
 | | Crystal Structure of NTB-A | | Descriptor: | CALCIUM ION, CHLORIDE ION, SLAM family member 6 | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2006-09-20 | | Release date: | 2006-10-17 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | NTB-A Receptor Crystal Structure: Insights into Homophilic Interactions in the Signaling Lymphocytic Activation Molecule Receptor Family.
Immunity, 25, 2006
|
|
6E25
 
 | |
8CU7
 
 | | Crystal structure of A2AAR-StaR2-bRIL in complex with a novel A2a antagonist, LJ-4517 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3R,4R)-2-[(8P)-6-amino-2-(hex-1-yn-1-yl)-8-(thiophen-2-yl)-9H-purin-9-yl]oxolane-3,4-diol, Adenosine receptor A2a,Soluble cytochrome b562, ... | | Authors: | Shiriaeva, A, Park, D.-J, Kim, G, Lee, Y, Hou, X, Jarhad, D.B, Kim, G, Yu, J, Hyun, Y.E, Kim, W, Gao, Z.-G, Jacobson, K.A, Han, G.W, Stevens, R.C, Jeong, L.S, Choi, S, Cherezov, V. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GPCR Agonist-to-Antagonist Conversion: Enabling the Design of Nucleoside Functional Switches for the A 2A Adenosine Receptor.
J.Med.Chem., 65, 2022
|
|
2TCI
 
 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | THIOCYANATE INSULIN, THIOCYANATE ION, ZINC ION | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
4E1C
 
 | | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ADP and Mg++ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-03-06 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and functional characterization of the Vibrio cholerae toxin
from the VgrG/MARTX family.
J.Biol.Chem., 2012
|
|
2IC3
 
 | |
7B1J
 
 | |
8D9M
 
 | | Cryo-EM of the OmcZ nanowires from Geobacter sulfurreducens | | Descriptor: | Cytochrome c, HEME C | | Authors: | Wang, F, Chan, C.H, Mustafa, K, Hochbaum, A.I, Bond, D.R, Egelman, E.H. | | Deposit date: | 2022-06-10 | | Release date: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of Geobacter OmcZ filaments suggests extracellular cytochrome polymers evolved independently multiple times.
Elife, 11, 2022
|
|
5OJJ
 
 | | Crystal structure of the Zn-bound ubiquitin-conjugating enzyme Ube2T | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Ubiquitin-conjugating enzyme E2 T, ... | | Authors: | Morreale, F.E, Testa, A, Chaugule, V.K, Bortoluzzi, A, Ciulli, A, Walden, H. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mind the Metal: A Fragment Library-Derived Zinc Impurity Binds the E2 Ubiquitin-Conjugating Enzyme Ube2T and Induces Structural Rearrangements.
J. Med. Chem., 60, 2017
|
|
8DEW
 
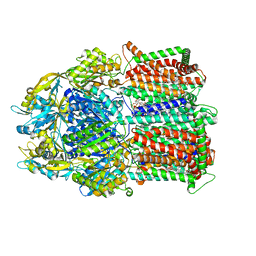 | |
4DD8
 
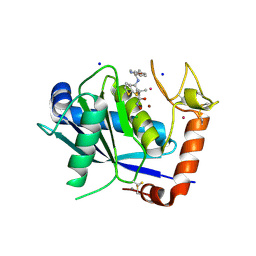 | | ADAM-8 metalloproteinase domain with bound batimastat | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hall, T, Shieh, H.S, Day, J.E, Caspers, N, Chrencik, J.E, Williams, J.M, Pegg, L.E, Pauley, A.M, Moon, A.F, Krahn, J.M, Fischer, D.H, Kiefer, J.R, Tomasselli, A.G, Zack, M.D. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human ADAM-8 catalytic domain complexed with batimastat.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1RZV
 
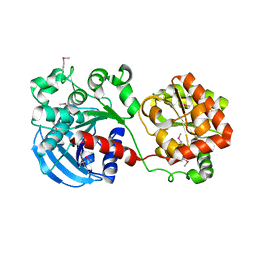 | | Crystal structure of the glycogen synthase from Agrobacterium tumefaciens (non-complexed form) | | Descriptor: | Glycogen synthase 1 | | Authors: | Buschiazzo, A, Guerin, M.E, Ugalde, J.E, Ugalde, R.A, Shepard, W, Alzari, P.M. | | Deposit date: | 2003-12-29 | | Release date: | 2004-08-31 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glycogen synthase: homologous enzymes catalyze glycogen synthesis and degradation.
Embo J., 23, 2004
|
|
6V0S
 
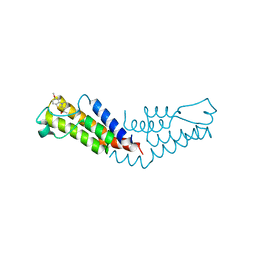 | |
7KMT
 
 | | Structure of the yeast TRAPPIII-Ypt1(Rab1) complex | | Descriptor: | GTP-binding protein YPT1, PALMITIC ACID, Trafficking protein particle complex III-specific subunit 85, ... | | Authors: | Joiner, A.M.N, Phillips, B.P, Miller, E.A, Fromme, J.C. | | Deposit date: | 2020-11-03 | | Release date: | 2021-06-02 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of TRAPPIII-mediated Rab1 activation.
Embo J., 40, 2021
|
|
2TRA
 
 | | RESTRAINED REFINEMENT OF TWO CRYSTALLINE FORMS OF YEAST ASPARTIC ACID AND PHENYLALANINE TRANSFER RNA CRYSTALS | | Descriptor: | MAGNESIUM ION, SPERMINE, TRNAASP | | Authors: | Westhof, E, Dumas, P, Moras, D. | | Deposit date: | 1987-11-06 | | Release date: | 1987-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals.
Acta Crystallogr.,Sect.A, 44, 1988
|
|
4I5O
 
 | | Crystal Structure of W-W-R ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.4787 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
1F29
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CRUZAIN BOUND TO A VINYL SULFONE DERIVED INHIBITOR (I) | | Descriptor: | 3-[[N-[MORPHOLIN-N-YL]-CARBONYL]-PHENYLALANINYL-AMINO]-5- PHENYL-PENTANE-1-SULFONYLBENZENE, CRUZAIN | | Authors: | Brinen, L.S, Hansell, E, Roush, W.R, McKerrow, J.H, Fletterick, R.J. | | Deposit date: | 2000-05-23 | | Release date: | 2000-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A target within the target: probing cruzain's P1' site to define structural determinants for the Chagas' disease protease.
Structure Fold.Des., 8, 2000
|
|
8DEU
 
 | |
4I6R
 
 | |
