1ZFJ
 
 | | INOSINE MONOPHOSPHATE DEHYDROGENASE (IMPDH; EC 1.1.1.205) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | INOSINE MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID | | Authors: | Zhang, R, Evans, G, Rotella, F.J, Westbrook, E.M, Beno, D, Huberman, E, Joachimiak, A, Collart, F.R. | | Deposit date: | 1999-03-29 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characteristics and crystal structure of bacterial inosine-5'-monophosphate dehydrogenase.
Biochemistry, 38, 1999
|
|
1ZG9
 
 | | Crystal Structure of 5-{[amino(imino)methyl]amino}-2-(sulfanylmethyl)pentanoic acid Bound to Activated Porcine Pancreatic Carboxypeptidase B | | Descriptor: | 5-{[AMINO(IMINO)METHYL]AMINO}-2-(SULFANYLMETHYL)PENTANOIC ACID, ZINC ION, procarboxypeptidase B | | Authors: | Adler, M, Bryant, J, Buckman, B, Islam, I, Larsen, B, Finster, S, Kent, L, May, K, Mohan, R, Yuan, S, Whitlow, M. | | Deposit date: | 2005-04-20 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of potent thiol-based inhibitors bound to carboxypeptidase b.
Biochemistry, 44, 2005
|
|
1ZHO
 
 | | The structure of a ribosomal protein L1 in complex with mRNA | | Descriptor: | 50S ribosomal protein L1, POTASSIUM ION, mRNA | | Authors: | Nevskaya, N, Tishchenko, S, Volchkov, S, Kljashtorny, V, Nikonova, E, Nikonov, O, Nikulin, A, Kohrer, C, Piendl, W, Zimmermann, R, Stockley, P, Garber, M, Nikonov, S. | | Deposit date: | 2005-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New insights into the interaction of ribosomal protein L1 with RNA.
J.Mol.Biol., 355, 2006
|
|
1ZIO
 
 | | PHOSPHOTRANSFERASE | | Descriptor: | ADENYLATE KINASE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Berry, M.B, Phillips Jr, G.N. | | Deposit date: | 1996-06-07 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of Bacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A, and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+.
Proteins, 32, 1998
|
|
1ZJD
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with Kunitz Protease Inhibitor Domain of Protease Nexin II | | Descriptor: | Catalytic Domain of Coagulation Factor XI, Kunitz Protease Inhibitory Domain of Protease Nexin II | | Authors: | Jin, L, Navaneetham, D, Pandey, P, Strickler, J.E, Babine, R.E, Walsh, P.N, Abdel-Meguid, S.S. | | Deposit date: | 2005-04-28 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mutational Analyses of the Molecular Interactions between the Catalytic Domain of Factor XIa and the Kunitz Protease Inhibitor Domain of Protease Nexin 2
J.Biol.Chem., 280, 2005
|
|
1Z94
 
 | | X-Ray Crystal Structure of Protein CV1439 from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvR12. | | Descriptor: | conserved hypothetical protein | | Authors: | Kuzin, A, Vorobiev, S.M, Yong, W, Forouhar, F, Xio, R, Ma, L.-C, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-03-31 | | Release date: | 2005-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Hypothetical Protein CV1439 from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CVR12.
To be Published
|
|
1ZM1
 
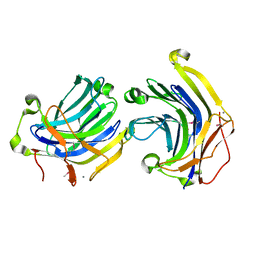 | | Crystal structures of complex F. succinogenes 1,3-1,4-beta-D-glucanase and beta-1,3-1,4-cellotriose | | Descriptor: | Beta-glucanase, CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Tsai, L.C, Shyur, L.F, Cheng, Y.S, Lee, S.H. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase in complex with beta-1,3-1,4-cellotriose
J.Mol.Biol., 354, 2005
|
|
1ZC6
 
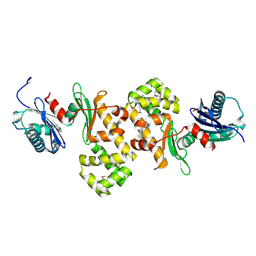 | | Crystal Structure of Putative N-acetylglucosamine Kinase from Chromobacterium violaceum. Northeast Structural Genomics Target Cvr23. | | Descriptor: | probable N-acetylglucosamine kinase | | Authors: | Vorobiev, S.M, Kuzin, A, Forouhar, F, Abashidze, M, Acton, T.B, Xiao, R, Ma, L.-C, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-11 | | Release date: | 2005-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Putative N-acetylglucosamine Kinase from Chromobacterium violaceum
To be Published
|
|
1Z6E
 
 | |
1ZA6
 
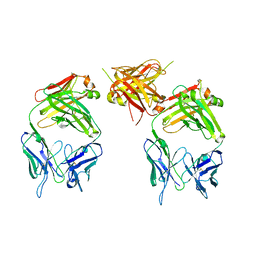 | | The structure of an antitumor CH2-domain-deleted humanized antibody | | Descriptor: | IGG Heavy chain, IGG Light chain | | Authors: | Larson, S.B, Day, J.S, Glaser, S, Braslawsky, G, McPherson, A. | | Deposit date: | 2005-04-05 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of an Antitumor C(H)2-domain-deleted Humanized Antibody.
J.Mol.Biol., 348, 2005
|
|
1Z9J
 
 | | Photosynthetic Reaction Center from Rhodobacter sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Thielges, M, Uyeda, G, Camara-Artigas, A, Kalman, L, Williams, J.C, Allen, J.P. | | Deposit date: | 2005-04-02 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Design of a Redox-Linked Active Metal Site: Manganese Bound to Bacterial Reaction Centers at a Site Resembling That of Photosystem II
Biochemistry, 44, 2005
|
|
1ZI6
 
 | | Crystal Structure Analysis of the dienelactone hydrolase (C123S) mutant- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
20GS
 
 | | GLUTATHIONE S-TRANSFERASE P1-1 COMPLEXED WITH CIBACRON BLUE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CIBACRON BLUE, GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Nuccetelli, M, Mazzetti, A.P, Parker, M.W. | | Deposit date: | 1997-12-16 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
1ZPT
 
 | | Escherichia coli Methylenetetrahydrofolate Reductase (reduced) complexed with NADH, pH 7.25 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 5,10-methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pejchal, R, Sargeant, R, Ludwig, M.L. | | Deposit date: | 2005-05-17 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of NADH and CH(3)-H(4)Folate Complexes of Escherichia coli Methylenetetrahydrofolate Reductase Reveal a Spartan Strategy for a Ping-Pong Reaction
Biochemistry, 44, 2005
|
|
1ZTJ
 
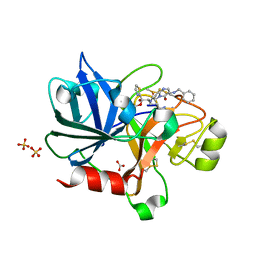 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with 2-(5-Benzylamino-2-methylsulfanyl-6-oxo-6H-pyrimidin-1-yl)-N-[4-guanidino-1-(thiazole-2-carbonyl)-butyl]-acetamide | | Descriptor: | 2-(5-BENZYLAMINO-2-METHYLSULFANYL-6-OXO-6H-PYRIMIDIN-1-YL)-N-[4-GUANIDINO-1-(THIAZOLE-2-CARBONYL)-BUTYL]-ACETAMIDE, BICARBONATE ION, Coagulation factor XI, ... | | Authors: | Nagafuji, P, Jin, L, Rynkiewicz, M, Quinn, J, Bibbins, F, Meyers, H, Babine, R.E, Strickler, J.E, Abdel-Meguid, S.S. | | Deposit date: | 2005-05-27 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pyrimidinone Inhibitors of a Thrombolytic Protease
To be Published
|
|
2E3C
 
 | |
1ZZB
 
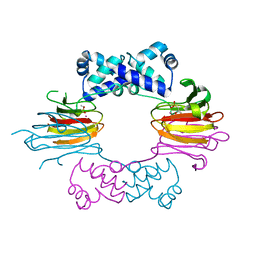 | | Crystal Structure of CoII HppE in Complex with Substrate | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, COBALT (II) ION, Hydroxypropylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZR
 
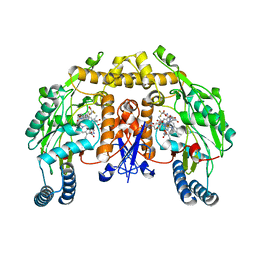 | | Rat nNOS D597N/M336V double mutant with L-N(omega)-Nitroarginine-(4R)-amino-L-proline amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, L-N(OMEGA)-NITROARGININE-(4R)-AMINO-L-PROLINE AMIDE, ... | | Authors: | Li, H, Flinspach, M.L, Igarashi, J, Jamal, J, Yang, W, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2005-06-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases.
Biochemistry, 44, 2005
|
|
1ZTD
 
 | | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus | | Descriptor: | Hypothetical Protein Pfu-631545-001 | | Authors: | Fu, Z.-Q, Horanyi, P, Florence, Q, Liu, Z.-J, Chen, L, Lee, D, Habel, J, Xu, H, Nguyen, D, Chang, S.-H, Zhou, W, Zhang, H, Jenney Jr, F.E, Sha, B, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-05-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus
To be Published
|
|
1ZTL
 
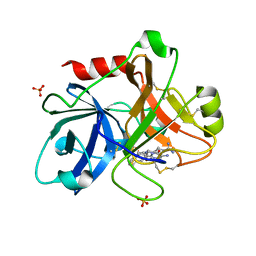 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with N-[4-Guanidino-1-(thiazole-2-carbonyl)-butyl]-2-{6-oxo-5-[(quinolin-8-ylmethyl)-amino]-2-m-tolyl-6H-pyrimidin-1-yl}-acetamide | | Descriptor: | Coagulation factor XI, N-[4-GUANIDINO-1-(THIAZOLE-2-CARBONYL)-BUTYL]-2-{6-OXO-5-[(QUINOLIN-8-YLMETHYL)-AMINO]-2-M-TOLYL-6H-PYRIMIDIN-1-YL}-ACETAMIDE, SULFATE ION | | Authors: | Nagafuji, P, Jin, L, Rynkiewicz, M, Quinn, J, Bibbins, F, Meyers, H, Babine, R, Strickler, J.E, Abdel-Meguid, S.S. | | Deposit date: | 2005-05-27 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pyrimidinone Inhibitors of a Thrombolytic Protease
To be Published
|
|
2EAQ
 
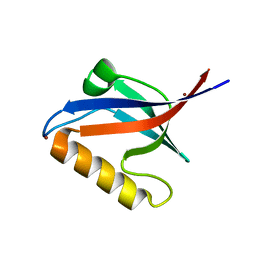 | | Crystal structure of PDZ domain of KIAA0858 (LIM), MS0793 from Homo sapiens | | Descriptor: | LIM domain only protein 7, NICKEL (II) ION | | Authors: | Xie, Y, Kishishita, S, Murayama, K, Takemoto, C, Shirozu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-31 | | Release date: | 2007-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal structure of PDZ domain of KIAA0858 (LIM), MS0793 from Homo sapiens
To be Published
|
|
1ZRN
 
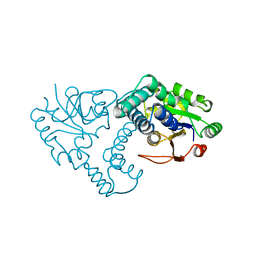 | | INTERMEDIATE STRUCTURE OF L-2-HALOACID DEHALOGENASE WITH MONOCHLOROACETATE | | Descriptor: | ACETIC ACID, L-2-HALOACID DEHALOGENASE | | Authors: | Li, Y.-F, Hata, Y, Fujii, T, Hisano, T, Nishihara, M, Kurihara, T, Esaki, N. | | Deposit date: | 1998-03-03 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of reaction intermediates of L-2-haloacid dehalogenase and implications for the reaction mechanism.
J.Biol.Chem., 273, 1998
|
|
1XVQ
 
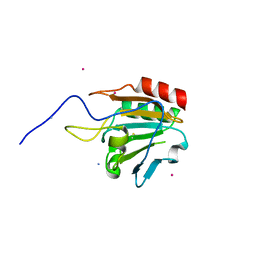 | | Crystal structure of thiol peroxidase from Mycobacterium tuberculosis | | Descriptor: | AMMONIUM ION, YTTRIUM (III) ION, thiol peroxidase | | Authors: | Rho, B.S, Pedelacq, J.D, Hung, L.W, Holton, J.M, Vigil, D, Kim, S.I, Park, M.S, Terwilliger, T.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-10-28 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and Structural Characterization of a Thiol Peroxidase from Mycobacterium tuberculosis.
J.Mol.Biol., 361, 2006
|
|
2DXE
 
 | | Crystal structure of nucleoside diphosphate kinase in complex with GDP | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-25 | | Release date: | 2007-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of nucleoside diphosphate kinase in complex with GDP
To be Published
|
|
1XXD
 
 | | Crystal Structure of the FXIa Catalytic Domain in Complex with mutated Ecotin | | Descriptor: | Coagulation factor XI, Ecotin | | Authors: | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
