3B55
 
 | | Crystal structure of the Q81BN2_BACCR protein from Bacillus cereus. NESG target BcR135 | | Descriptor: | CALCIUM ION, Succinoglycan biosynthesis protein | | Authors: | Vorobiev, S.M, Chen, Y, Kuzin, A.P, Seetharaman, J, Wang, H, Cunningham, K, Owens, L, Maglaqui, M, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Q81BN2_BACCR protein from Bacillus cereus.
To be Published
|
|
3BL2
 
 | | Crystal Structure of M11, the BCL-2 Homolog of Murine Gamma-herpesvirus 68, Complexed with Mouse Beclin1 (residues 106-124) | | Descriptor: | Beclin-1, V-bcl-2 | | Authors: | Oh, B.-H, Woo, J.-S, Ku, B. | | Deposit date: | 2007-12-10 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Bases for the Inhibition of Autophagy and Apoptosis by Viral BCL-2 of Murine gamma-Herpesvirus 68
Plos Pathog., 4, 2008
|
|
3AM6
 
 | |
3AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 Q9TQ44T | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1O9X
 
 | | HUMAN SERUM ALBUMIN COMPLEXED WITH TETRADECANOIC ACID (MYRISTIC ACID) AND HEMIN | | Descriptor: | MYRISTIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SERUM ALBUMIN | | Authors: | Zunszain, P.A, Ghuman, J, Komatsu, T, Tsuchida, E, Curry, S. | | Deposit date: | 2002-12-20 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure Analysis of Human Serum Albumin Complexed with Hemin and Fatty Acid
Bmc Struct.Biol., 3, 2003
|
|
3ANR
 
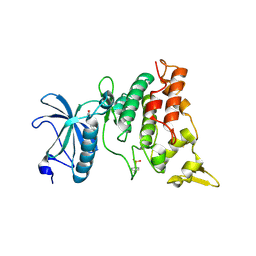 | | human DYRK1A/harmine complex | | Descriptor: | 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Nonaka, Y, Hosoya, T, Hagiwara, M, Ito, N. | | Deposit date: | 2010-09-06 | | Release date: | 2010-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of a novel selective inhibitor of the Down syndrome-related kinase Dyrk1A
Nat Commun, 1, 2010
|
|
3ANQ
 
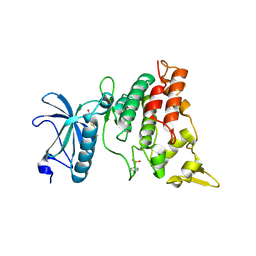 | | human DYRK1A/inhibitor complex | | Descriptor: | (1Z)-1-(3-ethyl-5-hydroxy-1,3-benzothiazol-2(3H)-ylidene)propan-2-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Nonaka, Y, Hosoya, T, Hagiwara, M, Ito, N. | | Deposit date: | 2010-09-06 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of a novel selective inhibitor of the Down syndrome-related kinase Dyrk1A
Nat Commun, 1, 2010
|
|
3AID
 
 | |
3AKY
 
 | |
3ATJ
 
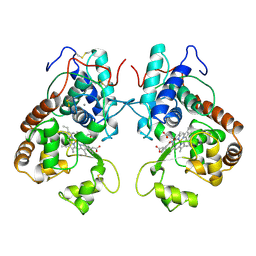 | | HEME LIGAND MUTANT OF RECOMBINANT HORSERADISH PEROXIDASE IN COMPLEX WITH BENZHYDROXAMIC ACID | | Descriptor: | BENZHYDROXAMIC ACID, CALCIUM ION, PROTEIN (HORSERADISH PEROXIDASE C1A), ... | | Authors: | Meno, K, White, C.G, Smith, A.T, Gajhede, M. | | Deposit date: | 1998-12-16 | | Release date: | 1999-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Catalytical Implications of a F221M Mutation in the Proximal Pocket of Horseradish Peroxidase C (HRP C)
To be Published
|
|
3AUJ
 
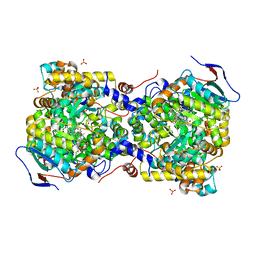 | | Structure of diol dehydratase complexed with glycerol | | Descriptor: | CALCIUM ION, COBALAMIN, Diol dehydrase alpha subunit, ... | | Authors: | Yamanishi, M, Kinoshita, K, Fukuoka, M, Shibata, T, Tobimatsu, T, Toraya, T. | | Deposit date: | 2011-02-07 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Redesign of coenzyme B(12) dependent diol dehydratase to be resistant to the mechanism-based inactivation by glycerol and act on longer chain 1,2-diols
Febs J., 279, 2012
|
|
3AQK
 
 | |
3AQS
 
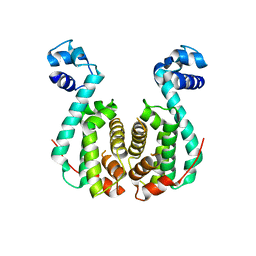 | | Crystal structure of RolR (NCGL1110) without ligand | | Descriptor: | Bacterial regulatory proteins, tetR family | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
3ARJ
 
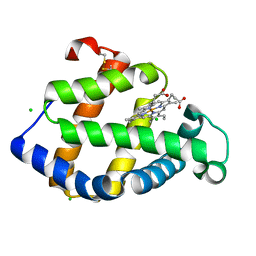 | | Cl- binding hemoglobin component V form Propsilocerus akamusi under 500 mM NaCl at pH 4.6 | | Descriptor: | CHLORIDE ION, Hemoglobin V, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kuwada, T, Hasegawa, T, Takagi, T, Shishikura, F. | | Deposit date: | 2010-12-02 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Involvement of the distal Arg residue in Cl- binding of midge larval haemoglobin
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3AU0
 
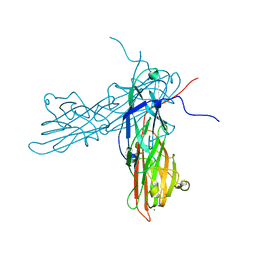 | | Structural and biochemical characterization of ClfB:ligand interactions | | Descriptor: | Clumping factor B, MAGNESIUM ION | | Authors: | Ganesh, V.K, Barbu, E.M, Deivanayagam, C.C.S, Le, B, Anderson, A.S, Matsuka, Y, Lin, S.L, Foster, T.F, Narayana, S.V.L, Hook, M. | | Deposit date: | 2011-01-28 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and biochemical characterization of ClfB:ligand interactions
To be published
|
|
3AU5
 
 | |
3AV3
 
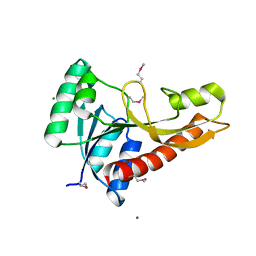 | | Crystal structure of glycinamide ribonucleotide transformylase 1 from Geobacillus kaustophilus | | Descriptor: | MAGNESIUM ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Kanagawa, M, Baba, S, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-02-18 | | Release date: | 2012-03-07 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures and reaction mechanisms of the two related enzymes, PurN and PurU.
J.Biochem., 154, 2013
|
|
3BCP
 
 | | Crystal Structure of The Swapped non covalent form of P19A/L28Q/N67D BS-RNase | | Descriptor: | Seminal ribonuclease | | Authors: | Merlino, A, Ercole, C, Picone, D, Pizzo, E, Mazzarella, L, Sica, F. | | Deposit date: | 2007-11-13 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The buried diversity of bovine seminal ribonuclease: shape and cytotoxicity of the swapped non-covalent form of the enzyme
J.Mol.Biol., 376, 2008
|
|
3B65
 
 | |
3BIJ
 
 | | Crystal structure of protein GSU0716 from Geobacter sulfurreducens. Northeast Structural Genomics target GsR13 | | Descriptor: | Uncharacterized protein GSU0716 | | Authors: | Forouhar, F, Neely, H, Su, M, Seetharaman, J, Benach, J, Conover, K, Fang, Y, Xiao, R, Owen, L.A, Maglaqui, M, Cunningham, K, Baran, M.C, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-11 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein GSU0716 from Geobacter sulfurreducens.
To be Published
|
|
3BAD
 
 | |
1Q0A
 
 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator (space group C222) | | Descriptor: | SULFATE ION, ZINC ION, Zn(II)-responsive regulator of zntA | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
3BGR
 
 | | Crystal structure of K103N/Y181C mutant HIV-1 reverse transcriptase (RT) in complex with TMC278 (Rilpivirine), a non-nucleoside RT inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Reverse transcriptase/ribonuclease H, ... | | Authors: | Das, K, Bauman, J.D, Clark Jr, A.D, Shatkin, A.J, Arnold, E. | | Deposit date: | 2007-11-27 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BC2
 
 | |
3BCC
 
 | | STIGMATELLIN AND ANTIMYCIN BOUND CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | ANTIMYCIN, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 1998-03-23 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Electron transfer by domain movement in cytochrome bc1.
Nature, 392, 1998
|
|
