3ZFF
 
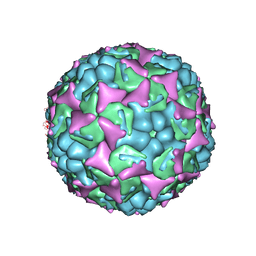 | | Human enterovirus 71 in complex with capsid binding inhibitor WIN51711 | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, VP1, VP2, ... | | Authors: | Plevka, P, Perera, R, Yap, M.L, Cardosa, J, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2012-12-11 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of Human Enterovirus 71 in Complex with a Capsid-Binding Inhibitor.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4E0P
 
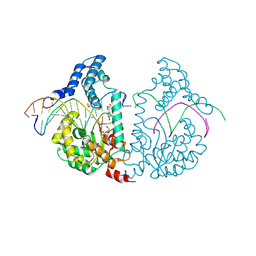 | | Protelomerase tela covalently complexed with substrate DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*TP*AP*AP*TP*AP*AP*CP*AP*AP*TP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*GP*AP*TP*AP*TP*TP*GP*TP*TP*AP*TP*TP*AP*TP*G)-3'), ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation.
Plos Biol., 11, 2013
|
|
4DTW
 
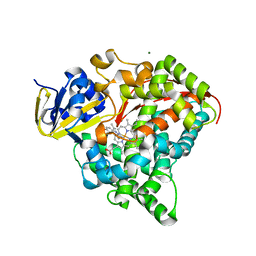 | | cytochrome P450 BM3h-8C8 MRI sensor bound to serotonin | | Descriptor: | Cytochrome P450 BM3 variant 8C8, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
4E0S
 
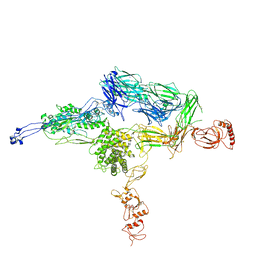 | | Crystal Structure of C5b-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C5, ... | | Authors: | Aleshin, A.E, Stec, B, DiScipio, R, Liddington, R.C. | | Deposit date: | 2012-03-05 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.21 Å) | | Cite: | Crystal structure of c5b-6 suggests structural basis for priming assembly of the membrane attack complex.
J.Biol.Chem., 287, 2012
|
|
4A2Q
 
 | | Structure of duck RIG-I tandem CARDs and helicase domain | | Descriptor: | RETINOIC ACID INDUCIBLE PROTEIN I | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-28 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for the Activation of Innate Immune Pattern Recognition Receptor Rig-I by Viral RNA.
Cell(Cambridge,Mass.), 147, 2011
|
|
4AB0
 
 | | X-ray crystal structure of Nicotiana alata defensin NaD1 | | Descriptor: | FLOWER-SPECIFIC DEFENSIN, PHOSPHATE ION | | Authors: | Mills, G.D, Lay, F.T, Hulett, M.D, Kvansakul, M. | | Deposit date: | 2011-12-06 | | Release date: | 2012-04-25 | | Last modified: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (1.636 Å) | | Cite: | Dimerization of Plant Defensin Nad1 Enhances its Antifungal Activity.
J.Biol.Chem., 287, 2012
|
|
4E0J
 
 | | Protelomerase tela R255A mutant complexed with DNA hairpin product | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*AP*TP*AP*AP*TP*AP*AP*CP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*CP*AP*TP*GP*AP*TP*AP*TP*TP*GP*TP*TP*AP*TP*TP*AP*TP*G)-3'), ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2012-03-04 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation.
Plos Biol., 11, 2013
|
|
6G7F
 
 | | Yeast 20S proteasome in complex with Cystargolide B | | Descriptor: | CHLORIDE ION, Cystargolide B- bound form, MAGNESIUM ION, ... | | Authors: | Groll, M, Tello-Aburto, R. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, synthesis, and evaluation of cystargolide-based beta-lactones as potent proteasome inhibitors.
Eur J Med Chem, 157, 2018
|
|
6FPA
 
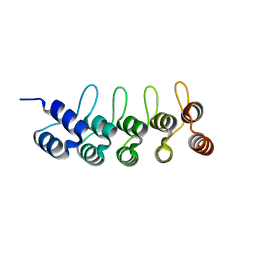 | | Crystal structure of anti-mTFP1 DARPin 1238_G01 in space group P212121 | | Descriptor: | DARPin 1238_G01 | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6GB7
 
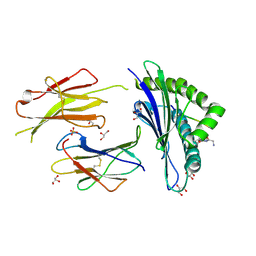 | | Structure of H-2Db with scoop loop from tapasin | | Descriptor: | Beta-2-microglobulin, GLY-GLY-LEU-SER, GLYCEROL, ... | | Authors: | Hafstrand, I, Sandalova, T, Achour, A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7DRC
 
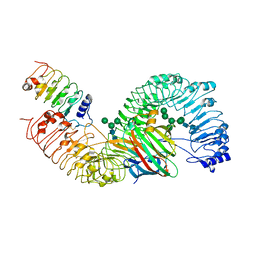 | | Cryo-EM structure of plant receptor like protein RXEG1 in complex with xyloglucanase XEG1 and BAK1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinosteroid insensitive 1-associated receptor kinase 1, ... | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2020-12-27 | | Release date: | 2022-06-22 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
3UME
 
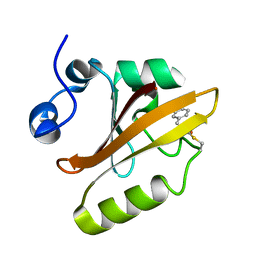 | | Structure of pB intermediate of Photoactive yellow protein (PYP) at pH 7 | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Tripathi, S, Srajer, V, Purwar, N, Henning, R, Schmidt, M. | | Deposit date: | 2011-11-13 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | pH Dependence of the Photoactive Yellow Protein Photocycle Investigated by Time-Resolved Crystallography.
Biophys.J., 102, 2012
|
|
4DHV
 
 | | Crystal structure of the Pyrococcus furiosus ferredoxin D14C variant containing the heterometallic [AgFe3S4] cluster | | Descriptor: | COBALT HEXAMMINE(III), Ferredoxin, SILVER/IRON/SULFUR CLUSTER | | Authors: | Jakab-Simon, I.N, Christensen, H.E.M, Haahr, L.T. | | Deposit date: | 2012-01-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Heterometallic [AgFe(3)S (4)] ferredoxin variants: synthesis, characterization, and the first crystal structure of an engineered heterometallic iron-sulfur protein.
J.Biol.Inorg.Chem., 18, 2013
|
|
6GK9
 
 | | Inhibited structure of IMPDH from Pseudomonas aeruginosa | | Descriptor: | (5~{S})-7-azanyl-5-(4-chlorophenyl)-2,4-bis(oxidanylidene)-1,5-dihydropyrano[2,3-d]pyrimidine-6-carbonitrile, Inosine-5'-monophosphate dehydrogenase, SULFATE ION | | Authors: | Labesse, G, Alexandre, T, Haouz, A, Munier-Lehmann, H. | | Deposit date: | 2018-05-18 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | First-in-class allosteric inhibitors of bacterial IMPDHs.
Eur J Med Chem, 167, 2019
|
|
6GOP
 
 | | Yeast 20S Proteasome in complex with Homosalinosporamide A | | Descriptor: | CHLORIDE ION, Homosalinosporamide A - bound form, MAGNESIUM ION, ... | | Authors: | Groll, M, Romo, D. | | Deposit date: | 2018-06-01 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | (-)-Homosalinosporamide A and Its Mode of Proteasome Inhibition: An X-ray Crystallographic Study.
Mar Drugs, 16, 2018
|
|
3UK6
 
 | | Crystal Structure of the Tip48 (Tip49b) hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 2 | | Authors: | Petukhov, M, Dagkessamanskaja, A, Bommer, M, Barrett, T, Tsaneva, I, Yakimov, A, Queval, R, Shvetsov, A, Khodorkovskiy, M, Kas, E, Grigoriev, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Large-Scale Conformational Flexibility Determines the Properties of AAA+ TIP49 ATPases.
Structure, 20, 2012
|
|
3UMG
 
 | | Crystal Structure of the Defluorinating L-2-Haloacid Dehalogenase Rha0230 | | Descriptor: | CHLORIDE ION, Haloacid dehalogenase | | Authors: | Chan, P.W.Y, Savchenko, A, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-11-13 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural adaptations of L-2-haloacid dehalogenases that enable hydrolytic defluorination
To be Published
|
|
3UNB
 
 | | Mouse constitutive 20S proteasome in complex with PR-957 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
4A34
 
 | |
4A5W
 
 | | Crystal structure of C5b6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COMPLEMENT C5, ... | | Authors: | Hadders, M.A, Bubeck, D, Forneris, F, Pangburn, M, Llorca, O, Lea, S.M, Gros, P. | | Deposit date: | 2011-10-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Assembly and Regulation of the Membrane Attack Complex Based on Structures of C5B6 and Sc5B9.
Cell Rep., 1, 2012
|
|
4DQF
 
 | | Crystal Structure of (G16A/L38A) HIV-1 Protease in Complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, 1,2-ETHANEDIOL, Aspartyl protease, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-02-15 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
4DIP
 
 | | Crystal structure of human Peptidyl-prolyl cis-trans isomerase FKBP14 | | Descriptor: | PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase FKBP14, SODIUM ION | | Authors: | Krojer, T, Kiyani, W, Goubin, S, Muniz, J.R.C, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Oppermann, U, Zschocke, J, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-01-31 | | Release date: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of human Peptidyl-prolyl cis-trans isomerase FKBP14
To be Published
|
|
4DTY
 
 | | cytochrome P450 BM3h-8C8 MRI sensor, no ligand | | Descriptor: | MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 variant 8C8 | | Authors: | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
436D
 
 | |
4DQG
 
 | |
