7FFA
 
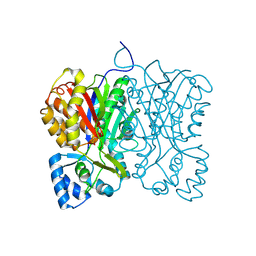 | | Diarylpentanoid-producing polyketide synthase from Aquilaria sinensis | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Kodama, T, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
7F8L
 
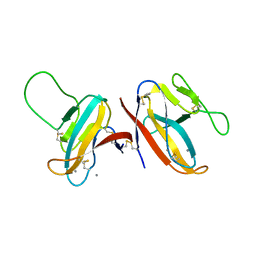 | |
7F8S
 
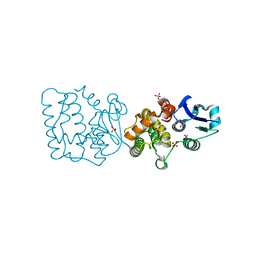 | | Pennisetum glaucum (Pearl millet) dehydroascorbate reductase (DHAR) with catalytic cysteine (Cy20) in sulphenic and sulfinic acid forms. | | Descriptor: | Dehydroascorbate reductase, SULFATE ION | | Authors: | Das, B.K, Kumar, A, Sreeshma, N.S, Arockiasamy, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Comparative kinetic analysis of ascorbate (Vitamin-C) recycling dehydroascorbate reductases from plants and humans.
Biochem.Biophys.Res.Commun., 591, 2021
|
|
7ER7
 
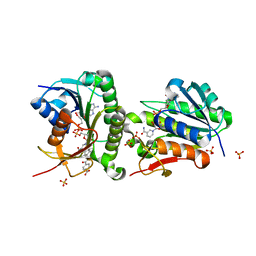 | | Crystal structure of hyman Biliverdin IX-beta reductase B with Tamibarotene (A80) | | Descriptor: | 4-[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbamoyl]benzoic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7FFC
 
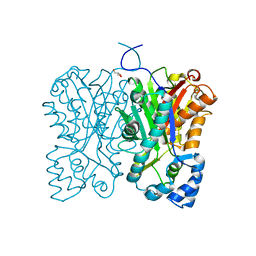 | | Diarylpentanoid-producing polyketide synthase (A210E mutant) | | Descriptor: | GLYCEROL, Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Kodama, T, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
7EQJ
 
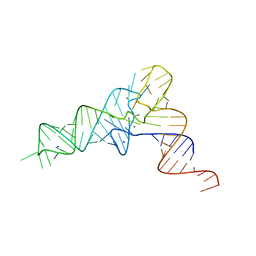 | | crystal structure of E. coli Valine tRNA | | Descriptor: | MAGNESIUM ION, RNA (76-MER), SODIUM ION | | Authors: | Kim, J, Jeong, H. | | Deposit date: | 2021-05-03 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Unique anticodon loop conformation with the flipped-out wobble nucleotide in the crystal structure of unbound tRNA Val .
Rna, 27, 2021
|
|
7FFH
 
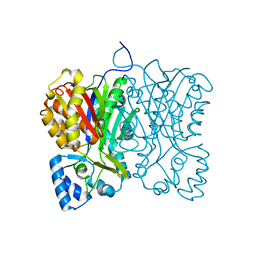 | | Diarylpentanoid-producing polyketide synthase (N199L mutant) | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Takeshi, K, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
7F81
 
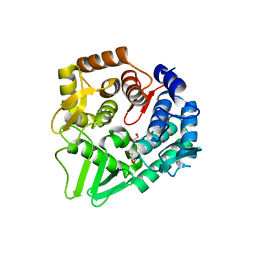 | | Structure of the bacterial cellulose synthase subunit Z from Enterobacter sp. CJF-002 | | Descriptor: | GLYCEROL, Glucanase, S,R MESO-TARTARIC ACID | | Authors: | Fujiwara, T, Fujishima, A, Yao, M. | | Deposit date: | 2021-06-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural snapshot of a glycoside hydrolase family 8 endo-beta-1,4-glucanase capturing the state after cleavage of the scissile bond.
Acta Crystallogr.,Sect.D, 78, 2022
|
|
7ERD
 
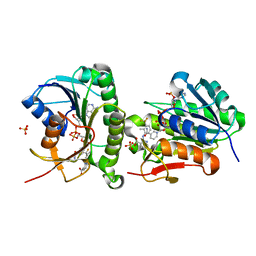 | | Crystal structure of human Biliverdin IX-beta reductase B with Flunixin Meglumin (FMG) | | Descriptor: | 2-[[2-methyl-3-(trifluoromethyl)phenyl]amino]pyridine-3-carboxylic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ER8
 
 | | Crystal structure of human Biliverdin IX-beta reductase B with Sulfasalazine (SAS) | | Descriptor: | 2-HYDROXY-(5-([4-(2-PYRIDINYLAMINO)SULFONYL]PHENYL)AZO)BENZOIC ACID, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7FCD
 
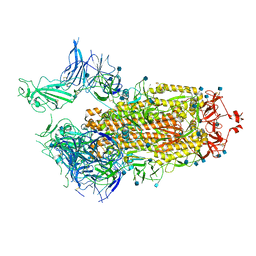 | | Structure of the SARS-CoV-2 A372T spike glycoprotein (open) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Loss of Spike N370 glycosylation as an important evolutionary event for the enhanced infectivity of SARS-CoV-2.
Cell Res., 32, 2022
|
|
7ERB
 
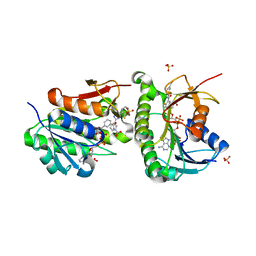 | | Crystal structure of human Biliverdin IX-beta reductase B with Ataluren (PTC) | | Descriptor: | 3-[5-(2-fluorophenyl)-1,2,4-oxadiazol-3-yl]benzoic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ESI
 
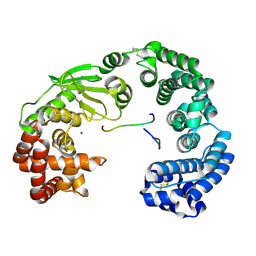 | | Crystal structure of the collagenase unit of a Vibrio collagenase from Vibrio harveyi VHJR7 at 1. 8 angstrom resolution. | | Descriptor: | CALCIUM ION, Collagenase unit (CU), Peptide P1, ... | | Authors: | Cao, H.Y, Wang, Y, Peng, M, Zhang, Y.Z. | | Deposit date: | 2021-05-11 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Vibrio collagenase VhaC provides insight into the mechanism of bacterial collagenolysis.
Nat Commun, 13, 2022
|
|
7FE1
 
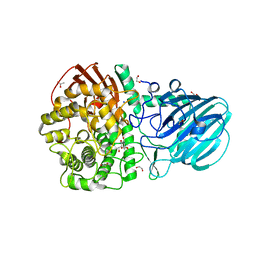 | | Crystal structure of GH92 alpha-1,2-mannosidase from Enterococcus faecalis ATCC 10100 in complex with methyl alpha-1,2-C-mannobioside | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Alpha-1,2-mannosidase, ... | | Authors: | Miyazaki, T, Alonso-Gil, S. | | Deposit date: | 2021-07-19 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Unlocking the Hydrolytic Mechanism of GH92 alpha-1,2-Mannosidases: Computation Inspires the use of C-Glycosides as Michaelis Complex Mimics.
Chemistry, 28, 2022
|
|
7F5F
 
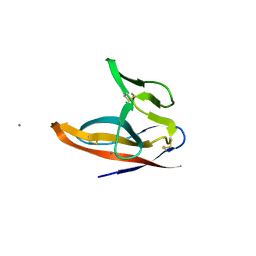 | | SARS-CoV-2 ORF8 S84 | | Descriptor: | CALCIUM ION, ORF8 protein | | Authors: | Chen, S, Zhou, Z, Chen, X. | | Deposit date: | 2021-06-22 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structures of Bat and Human Coronavirus ORF8 Protein Ig-Like Domain Provide Insights Into the Diversity of Immune Responses.
Front Immunol, 12, 2021
|
|
7FCZ
 
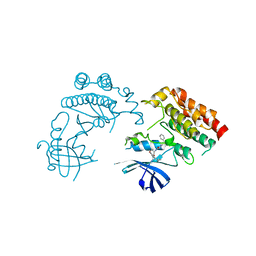 | | Crystal Structure of human RIPK1 kinase domain in complex with a novel inhibitor | | Descriptor: | N-[(3S)-7-(2-cyclopropylethynyl)-5-methyl-4-oxidanylidene-2,3-dihydro-1,5-benzoxazepin-3-yl]-5-(phenylmethyl)-4H-1,2,4-triazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Su, H.X, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Potent and Selective RIPK1 Inhibitors Targeting Dual-Pockets for the Treatment of Systemic Inflammatory Response Syndrome and Sepsis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7FD0
 
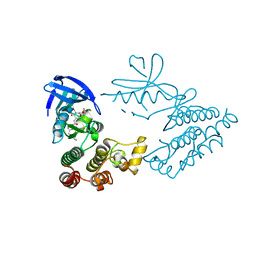 | | Crystal Structure of human RIPK1 kinase domain in complex with a novel inhibitor | | Descriptor: | N-[(3S)-5-methyl-7-[2-(oxan-4-yl)ethynyl]-4-oxidanylidene-2,3-dihydro-1,5-benzoxazepin-3-yl]-5-(phenylmethyl)-4H-1,2,4-triazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Su, H.X, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and Selective RIPK1 Inhibitors Targeting Dual-Pockets for the Treatment of Systemic Inflammatory Response Syndrome and Sepsis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7F8J
 
 | | Cryo-EM structure of human pannexin-1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7ERA
 
 | | Crystal structure of human Biliverdin IX-beta reductase B with Olsalazine Sodium (OSS) | | Descriptor: | 5-[(E)-(3-carboxy-4-oxidanyl-phenyl)diazenyl]-2-oxidanyl-benzoic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7F82
 
 | | Structure of the bacterial cellulose synthase subunit Z in complex with cellooligosaccharides from Enterobacter sp. CJF-002 | | Descriptor: | Glucanase, S,R MESO-TARTARIC ACID, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Fujiwara, T, Fujishima, A, Yao, M. | | Deposit date: | 2021-06-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural snapshot of a glycoside hydrolase family 8 endo-beta-1,4-glucanase capturing the state after cleavage of the scissile bond.
Acta Crystallogr.,Sect.D, 78, 2022
|
|
7EYK
 
 | |
7FC9
 
 | | Crystal structure of CmABCB1 in lipidic mesophase revealed by LCP-SFX | | Descriptor: | ACETATE ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Pan, D, Oyama, R, Sato, T, Nakane, T, Mizunuma, R, Matsuoka, K, Joti, Y, Tono, K, Nango, E, Iwata, S, Nakatsu, T, Kato, H. | | Deposit date: | 2021-07-14 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of CmABCB1 multi-drug exporter in lipidic mesophase revealed by LCP-SFX.
Iucrj, 9, 2022
|
|
7F8N
 
 | | Human pannexin-1 showing a conformational change in the N-terminal domain and blocked pore | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7FE2
 
 | | Crystal structure of the mutant E494Q of GH92 alpha-1,2-mannosidase from Enterococcus faecalis ATCC 10100 in complex with alpha-1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Alpha-1,2-mannosidase, ... | | Authors: | Miyazaki, T, Alonso-Gil, S. | | Deposit date: | 2021-07-19 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unlocking the Hydrolytic Mechanism of GH92 alpha-1,2-Mannosidases: Computation Inspires the use of C-Glycosides as Michaelis Complex Mimics.
Chemistry, 28, 2022
|
|
7FCE
 
 | | Structure of the SARS-CoV-2 A372T spike glycoprotein (closed) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Loss of Spike N370 glycosylation as an important evolutionary event for the enhanced infectivity of SARS-CoV-2.
Cell Res., 32, 2022
|
|
