3FH6
 
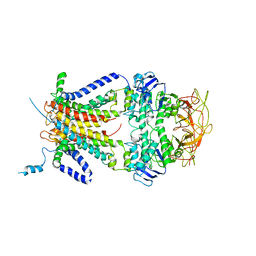 | | Crystal structure of the resting state maltose transporter from E. coli | | Descriptor: | Maltose transport system permease protein malF, Maltose transport system permease protein malG, Maltose/maltodextrin import ATP-binding protein malK | | Authors: | Khare, D, Oldham, M.L, Orelle, C, Davidson, A.L, Chen, J. | | Deposit date: | 2008-12-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Alternating access in maltose transporter mediated by rigid-body rotations.
Mol.Cell, 33, 2009
|
|
3FHT
 
 | |
3OYK
 
 | | Crystal structure of the PFV S217H mutant intasome bound to manganese | | Descriptor: | AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OM3
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with K362M mutation in the reduced state | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-02-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3F40
 
 | |
3OZI
 
 | | Crystal structure of the TIR domain from the flax disease resistance protein L6 | | Descriptor: | COBALT (II) ION, L6tr | | Authors: | Ve, T, Bernoux, M, Williams, S, Valkov, E, Warren, C, Hatters, D, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2010-09-25 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Analysis of a Plant Resistance Protein TIR Domain Reveals Interfaces for Self-Association, Signaling, and Autoregulation.
Cell Host Microbe, 9, 2011
|
|
3OMG
 
 | | Structure of human SND1 extended tudor domain in complex with the symmetrically dimethylated arginine PIWIL1 peptide R14me2s | | Descriptor: | Staphylococcal nuclease domain-containing protein 1, dimethylated arginine peptide R14me2s | | Authors: | Lam, R, Liu, K, Guo, Y.H, Bian, C.B, Xu, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3FJX
 
 | | E. coli EPSP synthase (T97I) liganded with S3P | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, SHIKIMATE-3-PHOSPHATE | | Authors: | Schonbrunn, E. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Glyphosate Resistance Resulting from the Double Mutation Thr97 -> Ile and Pro101 -> Ser in 5-Enolpyruvylshikimate-3-phosphate Synthase from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3FLE
 
 | | SE_1780 protein of unknown function from Staphylococcus epidermidis. | | Descriptor: | SE_1780 protein | | Authors: | Osipiuk, J, Hatzos, C, Clancy, S, Kim, Y, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | X-ray crystal structure of SE_1780 protein of unknown function from Staphylococcus epidermidis.
To be Published
|
|
3ONM
 
 | | Effector binding Domain of LysR-Type transcription factor RovM from Y. pseudotuberculosis | | Descriptor: | Transcriptional regulator LrhA | | Authors: | Quade, N, Diekmann, M, Haffke, M, Heroven, A.K, Dersch, P, Heinz, D.W. | | Deposit date: | 2010-08-30 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the effector-binding domain of the LysR-type transcription factor RovM from Yersinia pseudotuberculosis.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3F4S
 
 | |
3P10
 
 | |
3FLP
 
 | |
3FMZ
 
 | |
3OO8
 
 | |
3P2E
 
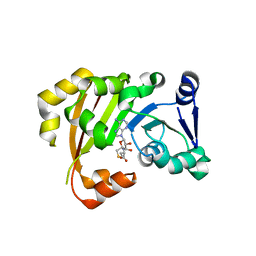 | | Structure of an antibiotic related Methyltransferase | | Descriptor: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sivaraman, J, Husain, N. | | Deposit date: | 2010-10-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for the methylation of A1408 in 16S rRNA by a panaminoglycoside resistance methyltransferase NpmA from a clinical isolate and analysis of the NpmA interactions with the 30S ribosomal subunit.
Nucleic Acids Res., 2010
|
|
3F6H
 
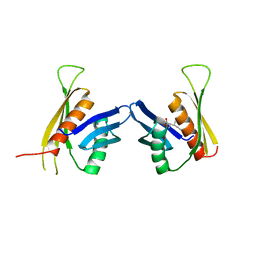 | | Crystal structure of the regulatory domain of LiCMS in complexed with isoleucine - type III | | Descriptor: | Alpha-isopropylmalate synthase, ISOLEUCINE, ZINC ION | | Authors: | Zhang, P, Ma, J, Zhao, G, Ding, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of the inhibitor selectivity and insights into the feedback inhibition mechanism of citramalate synthase from Leptospira interrogans
Biochem.J., 421, 2009
|
|
3F43
 
 | |
3F4T
 
 | |
3P3T
 
 | |
3F6U
 
 | | Crystal structure of human Activated Protein C (APC) complexed with PPACK | | Descriptor: | CALCIUM ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, ... | | Authors: | Schmidt, A.E, Padmanabhan, K, Underwood, M.C, Bode, W, Mather, T, Bajaj, S.P. | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermodynamic linkage between the S1 site, the Na+ site, and the Ca2+ site in the protease domain of human activated protein C (APC).
J.Biol.Chem., 277, 2002
|
|
3OQK
 
 | | Crystal Structure Analysis of Renin-indole-piperazin inhibitor complexes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-phenoxy-1-phenyl-3-(piperazin-1-ylcarbonyl)-1H-indole, GLYCEROL, ... | | Authors: | Bocskei, Z. | | Deposit date: | 2010-09-03 | | Release date: | 2010-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and optimization of a new class of potent and non-chiral indole-3-carboxamide-based renin inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3P45
 
 | | Crystal structure of apo-caspase-6 at physiological pH | | Descriptor: | caspase-6 | | Authors: | Mueller, I, Lamers, M.B.A.C, Ritchie, A.J, Dominguez, C, Munoz, I, Maillard, M, Kiselyov, A. | | Deposit date: | 2010-10-06 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structures of active and inhibitor-bound human Casp6
To be Published
|
|
3F79
 
 | |
3P5A
 
 | |
