4LGA
 
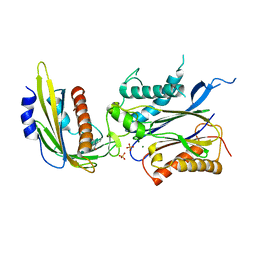 | | ABA-mimicking ligand N-(2-OXO-1-PROPYL-1,2,3,4-TETRAHYDROQUINOLIN-6-YL)-1-PHENYLMETHANESULFONAMIDE in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)-1-phenylmethanesulfonamide, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
4LGD
 
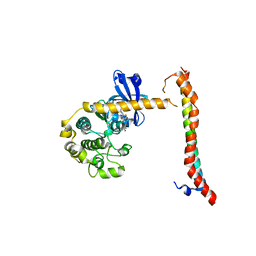 | | Structural Basis for Autoactivation of Human Mst2 Kinase and Its Regulation by RASSF5 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ras association domain family member 5, ... | | Authors: | Luo, X, Ni, L, Tomchick, D.R. | | Deposit date: | 2013-06-27 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Basis for Autoactivation of Human Mst2 Kinase and Its Regulation by RASSF5.
Structure, 21, 2013
|
|
4LGP
 
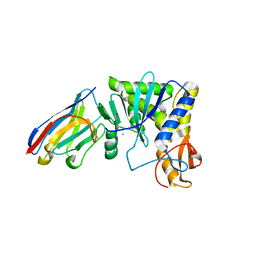 | | Ricin A chain bound to camelid nanobody (VHH1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ricin, ... | | Authors: | Rudolph, M.J, Cheung, J, Franklin, M, Burshteyn, F, Cassidy, M, Gary, E, Mantis, N. | | Deposit date: | 2013-06-28 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Ricin Toxin's Enzymatic Subunit (RTA) in Complex with Neutralizing and Non-Neutralizing Single-Chain Antibodies.
J.Mol.Biol., 426, 2014
|
|
4LHQ
 
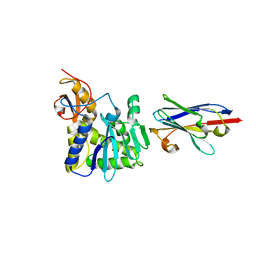 | | Ricin A chain bound to camelid nanobody (VHH8) | | Descriptor: | Camelid nanobody, Ricin | | Authors: | Rudolph, M.J, Cheung, J, Franklin, M, Burshteyn, F, Cassidy, M, Gary, E, Mantis, N. | | Deposit date: | 2013-07-01 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ricin Toxin's Enzymatic Subunit (RTA) in Complex with Neutralizing and Non-Neutralizing Single-Chain Antibodies.
J.Mol.Biol., 426, 2014
|
|
4LML
 
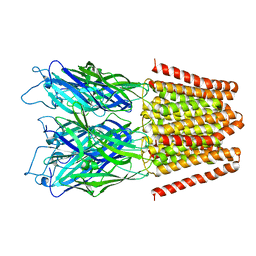 | | GLIC double mutant I9'A T25'A | | Descriptor: | Proton-gated ion channel | | Authors: | Grosman, C, Gonzalez-Gutierrez, G. | | Deposit date: | 2013-07-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LRY
 
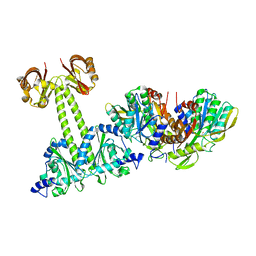 | | Crystal Structure of the E.coli DhaR(N)-DhaK(T79L) complex | | Descriptor: | GLYCEROL, PTS-dependent dihydroxyacetone kinase operon regulatory protein, PTS-dependent dihydroxyacetone kinase, ... | | Authors: | Shi, R, McDonald, L, Cygler, M, Ekiel, I. | | Deposit date: | 2013-07-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Coiled-Coil Helix Rotation Selects Repressing or Activating State of Transcriptional Regulator DhaR.
Structure, 22, 2014
|
|
4LV0
 
 | |
4LXG
 
 | |
9B1Q
 
 | | Crystal Structure of human Tryptophan 2,3-dioxygenase in complex with PYN1 inhibitor | | Descriptor: | (5P)-1-[(imidazolidin-1-yl)methyl]-5-(1H-indol-3-yl)-1H-1,2,3-benzotriazole, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase, ... | | Authors: | Geeraerts, Z, Yeh, S.-R. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.624 Å) | | Cite: | Structural Insights into Protein-Inhibitor Interactions in Human Tryptophan Dioxygenase.
J.Med.Chem., 67, 2024
|
|
4L78
 
 | | Xenon Trapping and Statistical Coupling Analysis Uncover Regions Important for Structure and Function of Multidomain Protein StPurL | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tanwar, A.S, Goyal, V.D, Choudhary, D, Panjikar, S, Anand, R. | | Deposit date: | 2013-06-13 | | Release date: | 2013-12-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Importance of hydrophobic cavities in allosteric regulation of formylglycinamide synthetase: insight from xenon trapping and statistical coupling analysis
Plos One, 8, 2013
|
|
4LCC
 
 | | Crystal structure of a human MAIT TCR in complex with a bacterial antigen bound to humanized bovine MR1 | | Descriptor: | 1-deoxy-1-[6-(hydroxymethyl)-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl]-D-arabinitol, Beta-2-microglobulin, MHC class I-related protein, ... | | Authors: | Lopez-Sagaseta, J, Adams, E.J. | | Deposit date: | 2013-06-21 | | Release date: | 2013-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.263 Å) | | Cite: | MAIT Recognition of a Stimulatory Bacterial Antigen Bound to MR1.
J.Immunol., 191, 2013
|
|
4WEF
 
 | | Structure of the Hemagglutinin-neuraminidase from Human parainfluenza virus type III: complex with difluorosialic acid | | Descriptor: | (2R,3R,4R,5R,6R)-5-acetamido-2,3-difluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]tetrahydro-2H-pyran-2-carboxylic acid, (3R,4R,5R,6R)-5-(acetylamino)-3-fluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]-3,4,5,6-tetrahydropyranium-2-carboxylate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Pilling, P, Barrett, S, McKimm-Breschkin, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic mechanism and novel receptor binding sites of human parainfluenza virus type 3 hemagglutinin-neuraminidase (hPIV3 HN)
Antiviral Res., 123, 2015
|
|
4V7O
 
 | | Proteasome Activator Complex | | Descriptor: | Proteasome activator BLM10, Proteasome component C1, Proteasome component C11, ... | | Authors: | Hill, C.P, Whitby, F.G. | | Deposit date: | 2009-12-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening.
Mol.Cell, 37, 2010
|
|
4LHI
 
 | |
4LJX
 
 | |
4LIZ
 
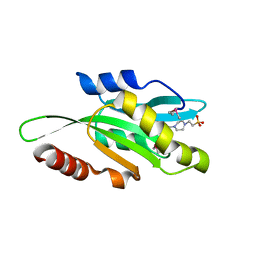 | | Crystal structure of coactosin from Entamoeba histolytica | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Actin-binding protein, cofilin/tropomyosin family protein, ... | | Authors: | Gourinath, S, Kumar, N. | | Deposit date: | 2013-07-04 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | EhCoactosin stabilizes actin filaments in the protist parasite Entamoeba histolytica.
Plos Pathog., 10, 2014
|
|
4LLG
 
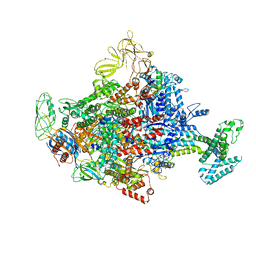 | | Crystal Structure Analysis of the E.coli holoenzyme/Gp2 complex | | Descriptor: | Bacterial RNA polymerase inhibitor, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2013-07-09 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7936 Å) | | Cite: | Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma 70 domain 1.1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LLL
 
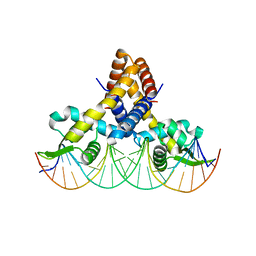 | | Crystal structure of S. aureus MepR-DNA complex | | Descriptor: | MepR, Palindromized mepR operator sequence | | Authors: | Birukou, I, Brennan, R.G. | | Deposit date: | 2013-07-09 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.036 Å) | | Cite: | Structural mechanism of transcription regulation of the Staphylococcus aureus multidrug efflux operon mepRA by the MarR family repressor MepR.
Nucleic Acids Res., 42, 2014
|
|
4LNY
 
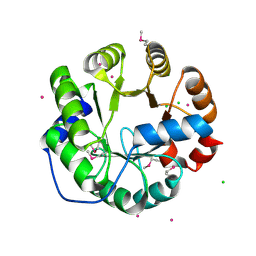 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR422 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Engineered Protein OR422 | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Maglaqui, M, Kogan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Crystal Structure of Engineered Protein OR422.
To be Published
|
|
4LMR
 
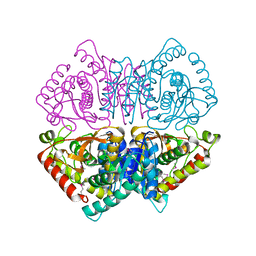 | | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579, NYSGRC Target 029452 | | Descriptor: | L-lactate dehydrogenase 1 | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-10 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579, NYSGRC Target 029452
To be Published
|
|
4L2Z
 
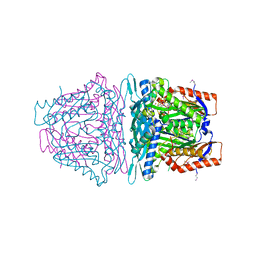 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus complexed with SAE and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-05 | | Release date: | 2013-06-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4L8S
 
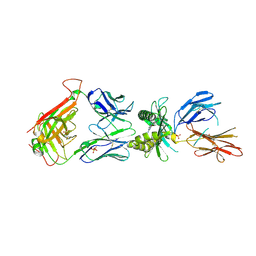 | |
4L9C
 
 | | Crystal structure of the FP domain of human F-box protein Fbxo7 (native) | | Descriptor: | F-box only protein 7, GLYCEROL | | Authors: | Du, Z, Huang, X, Shang, J, Yang, Y, Wang, G. | | Deposit date: | 2013-06-18 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the FP domain of Fbxo7 reveals a novel mode of protein-protein interaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4L9L
 
 | |
4LCH
 
 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-051 complex | | Descriptor: | (betaS)-Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N,beta-dihydroxy-beta-methyl-L-tyrosinamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
