4GL5
 
 | |
4FS3
 
 | | Crystal structure of Staphylococcus aureus enoyl-ACP reductase in complex with NADP and AFN-1252 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH] FabI, N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide, [[(2R,3S,4R,5R)-5-(3-aminocarbonyl-4H-pyridin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Kaplan, N, Yethon, J, Bardouniotis, E, Thalakada, R, Albert, M, Awrey, D.E, Romanov, V, Dorsey, M, Ramnauth, J, Clarke, T.E, Schmid, M.B, Berman, J, Pauls, H.W. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mode of Action, In Vitro Activity, and In Vivo Efficacy of AFN-1252, a Selective Antistaphylococcal FabI Inhibitor.
Antimicrob.Agents Chemother., 56, 2012
|
|
1X9E
 
 | | Crystal structure of HMG-CoA synthase from Enterococcus faecalis | | Descriptor: | HMG-CoA synthase, SULFATE ION | | Authors: | Steussy, C.N, Vartia, A.A, Burgner II, J.W, Sutherlin, A, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2004-08-20 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystal Structures of HMG-CoA Synthase from Enterococcus faecalis and a Complex with Its Second Substrate/Inhibitor Acetoacetyl-CoA
Biochemistry, 44, 2005
|
|
4FYJ
 
 | |
3LPE
 
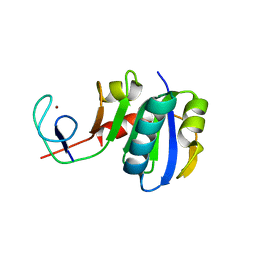 | | Crystal structure of Spt4/5NGN heterodimer complex from Methanococcus jannaschii | | Descriptor: | DNA-directed RNA polymerase subunit E'', Putative transcription antitermination protein nusG, ZINC ION | | Authors: | Hirtreiter, A, Damsma, G.E, Cheung, A.C.M, Klose, D, Grohmann, D, Vojnic, E, Martin, A.C.R, Cramer, P, Werner, F. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif.
Nucleic Acids Res., 38, 2010
|
|
1XEO
 
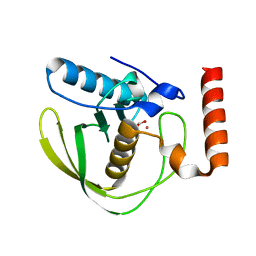 | | High Resolution Crystals Structure of Cobalt- Peptide Deformylase Bound To Formate | | Descriptor: | COBALT (II) ION, FORMIC ACID, Peptide deformylase | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
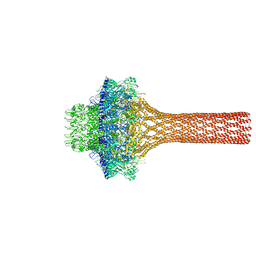
3LJ5
 
 | |
3LQ9
 
 | | Crystal structure of human REDD1, a hypoxia-induced regulator of mTOR | | Descriptor: | DNA-damage-inducible transcript 4 protein | | Authors: | Vega-Rubin-de-Celis, S, Abdallah, Z, Brugarolas, J, Zhang, X. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis and functional implications of the negative mTORC1 regulator REDD1.
Biochemistry, 49, 2010
|
|
1XFI
 
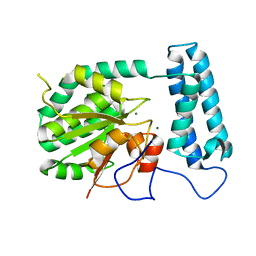 | | X-ray structure of gene product from Arabidopsis thaliana At2g17340 | | Descriptor: | MAGNESIUM ION, unknown protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-09-14 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure at 1.7 A resolution of the protein product of the At2g17340 gene from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
4G4S
 
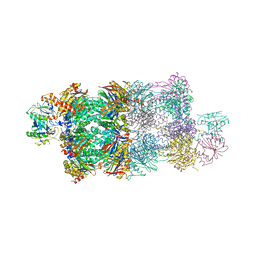 | | Structure of Proteasome-Pba1-Pba2 Complex | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-4-methyl-1-oxopentan-2-yl]-L-leucinamide, Proteasome assembly chaperone 2, ... | | Authors: | Kish-Trier, E, Robinson, H, Stadtmueller, B.M, Hill, C.P. | | Deposit date: | 2012-07-16 | | Release date: | 2012-09-05 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of a Proteasome Pba1-Pba2 Complex: IMPLICATIONS FOR PROTEASOME ASSEMBLY, ACTIVATION, AND BIOLOGICAL FUNCTION.
J.Biol.Chem., 287, 2012
|
|
3LU9
 
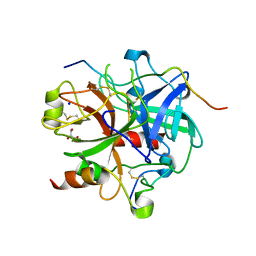 | | Crystal structure of human thrombin mutant S195A in complex with the extracellular fragment of human PAR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Proteinase-activated receptor 1, ... | | Authors: | Gandhi, P.S, Chen, Z, Di Cera, E. | | Deposit date: | 2010-02-17 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of thrombin bound to the uncleaved extracellular fragment of PAR1.
J.Biol.Chem., 285, 2010
|
|
1X9J
 
 | |
1X9F
 
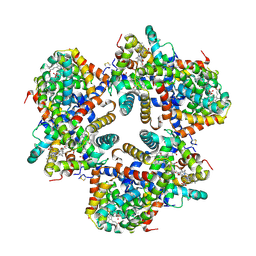 | | Hemoglobin Dodecamer from Lumbricus Erythrocruorin | | Descriptor: | CARBON MONOXIDE, Globin II, extracellular, ... | | Authors: | Strand, K, Knapp, J.E, Bhyravbhatla, B, Royer Jr, W.E. | | Deposit date: | 2004-08-20 | | Release date: | 2004-11-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the hemoglobin dodecamer from lumbricus erythrocruorin: allosteric core of giant annelid respiratory complexes
J.Mol.Biol., 344, 2004
|
|
3LYA
 
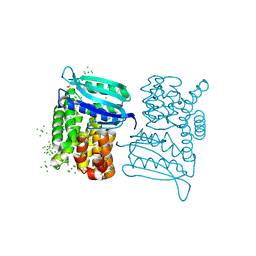 | |
4J72
 
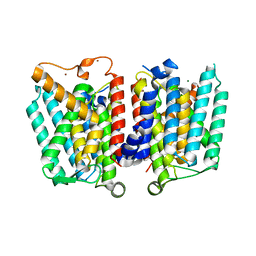 | | Crystal Structure of polyprenyl-phosphate N-acetyl hexosamine 1-phosphate transferase | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Lee, S.Y, Chung, B.C, Gillespie, R.A, Kwon, D.Y, Guan, Z, Zhou, P, Hong, J. | | Deposit date: | 2013-02-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis.
Science, 341, 2013
|
|
2AAZ
 
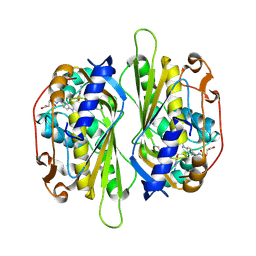 | | Cryptococcus neoformans thymidylate synthase complexed with substrate and an antifolate | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Finer-Moore, J.S, Anderson, A.C, O'Neil, R.H, Costi, M.P, Ferrari, S, Krucinski, J, Stroud, R.M. | | Deposit date: | 2005-07-14 | | Release date: | 2005-12-06 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The structure of Cryptococcus neoformans thymidylate synthase suggests strategies for using target dynamics for species-specific inhibition.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZZ5
 
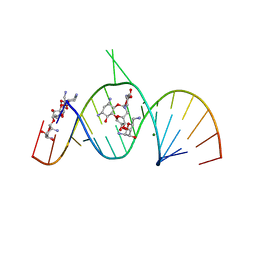 | | Molecular Recognition of RNA by Neomycin and a Restricted Neomycin Derivative | | Descriptor: | 13,15-DIAMINO-2-(AMINOMETHYL)-3,4,9,12-TETRAHYDROXYHEXADECAHYDRO-2H-7,10-EPOXYPYRANO[2,3-B][1,10,4]BENZODIOXAZACYCLODODECIN-8-YL 2,6-DIAMINO-2,6-DIDEOXYHEXOPYRANOSIDE, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*C)-3', 5'-R(*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*GP*G)-3', ... | | Authors: | Zhao, Q, Zhao, F, Blount, K.F, Han, Q, Tor, Y, Hermann, T. | | Deposit date: | 2005-06-13 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular recognition of RNA by neomycin and a restricted neomycin derivative
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
4RKU
 
 | | Crystal structure of plant Photosystem I at 3 Angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Mazor, Y, Borovikova, A, Greenberg, I, Nelson, N. | | Deposit date: | 2014-10-14 | | Release date: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of plant Photosystem I at 3.1 Angstrom resolution
To be Published
|
|
2A2R
 
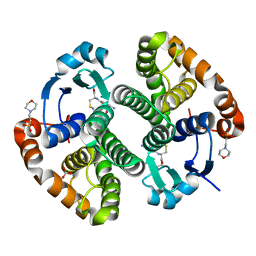 | | Crystal Structure of Glutathione Transferase Pi in complex with S-nitrosoglutathione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5-[1-(CARBOXYLATOMETHYLCARBAMOYL)-2-NITROSOSULFANYL-ETHYL]AMINO-5-OXO-PENTANOATE, CALCIUM ION, ... | | Authors: | Parker, L.J, Morton, C.J, Adams, J.J, Parker, M.W. | | Deposit date: | 2005-06-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Calorimetric and structural studies of the nitric oxide carrier S-nitrosoglutathione bound to human glutathione transferase P1-1
Protein Sci., 15, 2006
|
|
2AAE
 
 | | THE ROLE OF HISTIDINE-40 IN RIBONUCLEASE T1 CATALYSIS: THREE-DIMENSIONAL STRUCTURES OF THE PARTIALLY ACTIVE HIS40LYS MUTANT | | Descriptor: | CALCIUM ION, PHOSPHATE ION, RIBONUCLEASE T1 | | Authors: | Zegers, I, Verhelst, P, Choe, C.W, Steyaert, J, Heinemann, U, Wyns, L, Saenger, W. | | Deposit date: | 1992-09-15 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of histidine-40 in ribonuclease T1 catalysis: three-dimensionalstructures of the partially active His40Lys mutant.
Biochemistry, 31, 1992
|
|
4OIO
 
 | | Crystal structure of Thermus thermophilus pre-insertion substrate complex for de novo transcription initiation | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
2A46
 
 | |
2A63
 
 | |
2A6E
 
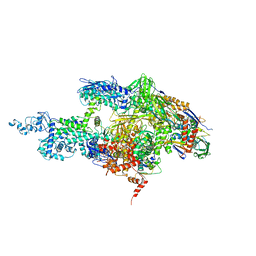 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
4I39
 
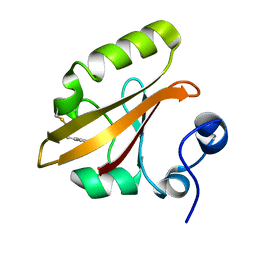 | | Structures of ICT and PR1 intermediates from time-resolved laue crystallography collected at 14ID-B, APS | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
