6PAU
 
 | |
8ONJ
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant R88L | | Descriptor: | Aminotransferase class IV, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
5KLC
 
 | | Structure of CBM_E1, a novel carbohydrate-binding module found by sugar cane soil metagenome | | Descriptor: | Carbohydrate binding module E1 | | Authors: | Liberato, M.V, Campos, B.M, Zeri, A.C.M, Squina, F.M. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | A Novel Carbohydrate-binding Module from Sugar Cane Soil Metagenome Featuring Unique Structural and Carbohydrate Affinity Properties.
J.Biol.Chem., 291, 2016
|
|
5HSW
 
 | | KSHV SOX RNA complex | | Descriptor: | ACETATE ION, ORF 37, RNA (5'-R(P*UP*CP*UP*UP*GP*AP*AP*GP*CP*AP*GP*CP*UP*UP*CP*CP*AP*G)-3') | | Authors: | Barrett, T.E. | | Deposit date: | 2016-01-26 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | KSHV SOX mediated host shutoff: the molecular mechanism underlying mRNA transcript processing.
Nucleic Acids Res., 45, 2017
|
|
5HQ0
 
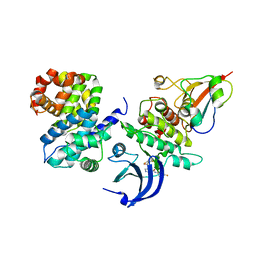 | | Ternary complex of human proteins CDK1, Cyclin B and CKS2, bound to an inhibitor | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Noble, M.E, Martin, M.P, Korolchuk, S, Brown, N.R, Moukhametzianov, R, Stanley, W.A. | | Deposit date: | 2016-01-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CDK1 structures reveal conserved and unique features of the essential cell cycle CDK.
Nat Commun, 6, 2015
|
|
6G9A
 
 | |
6VC7
 
 | |
8ORA
 
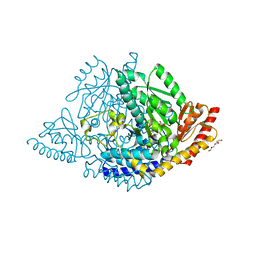 | | Human holo aromatic L-amino acid decarboxylase (AADC) external aldimine with L-Dopa methylester | | Descriptor: | Dopa decarboxylase (Aromatic L-amino acid decarboxylase), PYRIDOXAL-5'-PHOSPHATE, TETRAETHYLENE GLYCOL, ... | | Authors: | Bisello, G, Perduca, M, Bertoldi, M. | | Deposit date: | 2023-04-13 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human aromatic amino acid decarboxylase is an asymmetric and flexible enzyme: Implication in aromatic amino acid decarboxylase deficiency.
Protein Sci., 32, 2023
|
|
5KM9
 
 | |
5VZS
 
 | | BRD4-BD1 in complex with Cpd19 (3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-N-methyl-1-(tetrahydro-2H-pyran-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide) | | Descriptor: | 1,2-ETHANEDIOL, 3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1-(oxan-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, Bromodomain-containing protein 4 | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-29 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of CBP/P300
To be published
|
|
5HQL
 
 | | Structure function studies of R. palustris RubisCO (A47V-M331A mutant; CABP-bound; no expression tag) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure function studies of R. palustris RubisCO
To Be Published
|
|
5DXA
 
 | |
6Y8G
 
 | | selenomethionine derivative of ferulic acid esterase (FAE) | | Descriptor: | CADMIUM ION, Endo-1,4-beta-xylanase Y, GLYCEROL | | Authors: | von Stetten, D, Mueller-Dieckmann, C, Carpentier, P, Flot, D. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ID30A-3 (MASSIF-3) - a beamline for macromolecular crystallography at the ESRF with a small intense beam.
J.Synchrotron Radiat., 27, 2020
|
|
5KJY
 
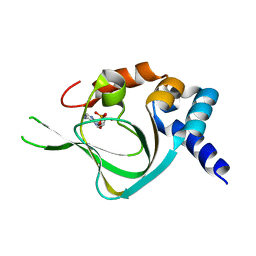 | | Co-crystal structure of PKA RI alpha CNB-B mutant (G316R/A336T) with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Lorenz, R, Moon, E, Kim, J.J, Huang, G.Y, Kim, C, Herberg, F.W. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of PKA cyclic nucleotide-binding domains reveal novel aspects of cyclic nucleotide selectivity.
Biochem. J., 474, 2017
|
|
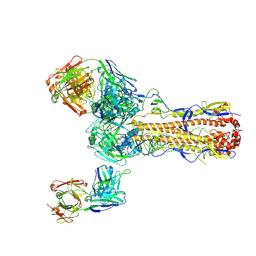
6PDX
 
 | |
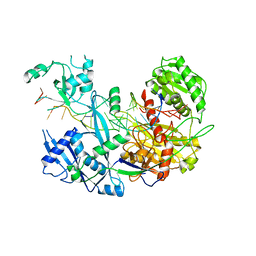
6N4O
 
 | |
8OWO
 
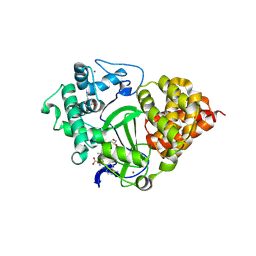 | | SMYD3 in complex with fragment FL01507 | | Descriptor: | 3-oxidanylbenzenecarbonitrile, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Lund, B.A, Cederfelt, D, Dobritzsch, D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of fragments targeting SMYD3 using highly sensitive kinetic and multiplexed biosensor-based screening.
Rsc Med Chem, 15, 2024
|
|
6Y8Y
 
 | | Structure of Baltic Herring (Clupea Harengus) Phosphoglucomutase 5 (PGM5) with bound Glucose-1-Phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Gustafsson, R, Eckhard, U, Selmer, M. | | Deposit date: | 2020-03-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Characterization of Phosphoglucomutase 5 from Atlantic and Baltic Herring-An Inactive Enzyme with Intact Substrate Binding.
Biomolecules, 10, 2020
|
|
6G9T
 
 | | CRYSTAL STRUCTURE OF CMY-136 class C BETA-LACTAMASE | | Descriptor: | Beta-lactamase, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Zavala, A, Retailleau, P, Naas, T, Iorga, B. | | Deposit date: | 2018-04-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Genetic, Biochemical, and Structural Characterization of CMY-136 beta-Lactamase, a Peculiar CMY-2 Variant.
Acs Infect Dis., 5, 2019
|
|
6N31
 
 | | WD repeats of human WDR12 | | Descriptor: | Ribosome biogenesis protein WDR12, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Zeng, H, Tempel, W, Li, Y, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | WD repeats of human WDR12
To Be Published
|
|
5DYU
 
 | | Crystal Structure of BAZ2B bromodomain in complex with fragment F39 | | Descriptor: | 1H-imidazol-5-ylmethanol, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Lolli, G, Caflisch, A. | | Deposit date: | 2015-09-25 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-Throughput Fragment Docking into the BAZ2B Bromodomain: Efficient in Silico Screening for X-Ray Crystallography.
Acs Chem.Biol., 11, 2016
|
|
5KK4
 
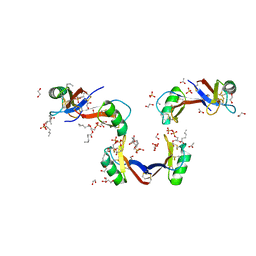 | | Crystal Structure of the Plant Defensin NsD7 bound to Phosphatidic Acid | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Kvansakul, M, Hulett, M.D, Lay, F.T. | | Deposit date: | 2016-06-21 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of phosphatidic acid by NsD7 mediates the formation of helical defensin-lipid oligomeric assemblies and membrane permeabilization.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6VDA
 
 | |
5KKJ
 
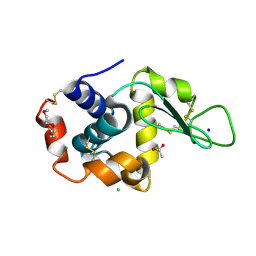 | | 2.0-Angstrom In situ Mylar structure of hen egg-white lysozyme (HEWL) at 293 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Broecker, J, Ernst, O.P. | | Deposit date: | 2016-06-21 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | A Versatile System for High-Throughput In Situ X-ray Screening and Data Collection of Soluble and Membrane-Protein Crystals.
Cryst Growth Des, 16, 2016
|
|
6PG4
 
 | |
