3EUF
 
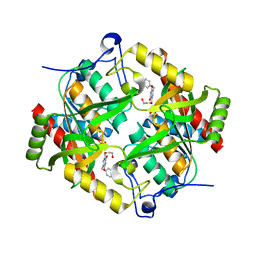 | | Crystal structure of BAU-bound human uridine phosphorylase 1 | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-BENZYLPYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, Uridine phosphorylase 1 | | Authors: | Roosild, T.P. | | Deposit date: | 2008-10-10 | | Release date: | 2009-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Implications of the structure of human uridine phosphorylase 1 on the development of novel inhibitors for improving the therapeutic window of fluoropyrimidine chemotherapy.
Bmc Struct.Biol., 9, 2009
|
|
7ML9
 
 | | The Mpp75Aa1.1 beta-pore-forming protein from Brevibacillus laterosporus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Insecticidal protein, ... | | Authors: | Rydel, T.J, Zheng, M, Evdokimov, A. | | Deposit date: | 2021-04-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional characterization of Mpp75Aa1.1, a putative beta-pore forming protein from Brevibacillus laterosporus active against the western corn rootworm.
Plos One, 16, 2021
|
|
3EL4
 
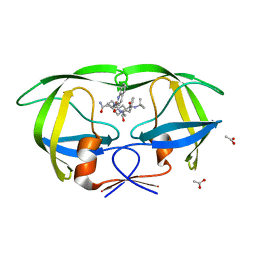 | | Crystal structure of inhibitor saquinavir (SQV) complexed with the multidrug HIV-1 protease variant L63P/V82T/I84V | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETATE ION, Protease | | Authors: | Prabu-Jeyabalan, M, King, N, Schiffer, C. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
7M1R
 
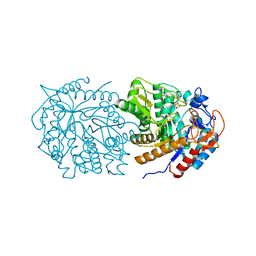 | |
7M2L
 
 | | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the alpha-and beta-site, sodium ion at the metal coordination site, and another F6F molecule at the enzyme beta-site at 1.60 Angstrom resolution. Two of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, CHLORIDE ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the alpha-and beta-site, sodium ion at the metal coordination site, and another F6F molecule at the enzyme beta-site at 1.60 Angstrom resolution.
To be Published
|
|
9MEA
 
 | |
6DGY
 
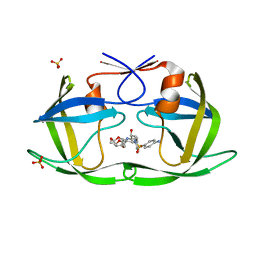 | | Crystal structure of HIV-1 Protease NL4-3 WT in complex with UMass1 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DH7
 
 | | Crystal structure of HIV-1 Protease NL4-3 I50V Mutant in complex with UMass1 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DH5
 
 | | Crystal structure of HIV-1 Protease NL4-3 V82I Mutant in complex with UMass6 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
5EI2
 
 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | Dual specificity protein kinase TTK, ~{N}-(2,4-dimethoxyphenyl)-8-(1-methylpyrazol-4-yl)pyrido[3,4-d]pyrimidin-2-amine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-29 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle Kinase 1 (MPS1) Using a Structure-Based Hybridization Approach.
J.Med.Chem., 59, 2016
|
|
7M3S
 
 | | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the alpha-and beta-site, sodium ion at the metal coordination site, and another F6F molecule at the enzyme beta-site at 1.55 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, CHLORIDE ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2021-03-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the alpha-and beta-site, sodium ion at the metal coordination site, and another F6F molecule at the enzyme beta-site at 1.55 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring.
To be Published
|
|
6DGZ
 
 | | Crystal structure of HIV-1 Protease NL4-3 WT in complex with UMass6 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
7WPT
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a fragment M2-80 and ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
9E0A
 
 | | Structure of proline utilization A complexed with 1,4-benzenedimethanol | | Descriptor: | (1,4-phenylene)dimethanol, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2024-10-17 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystallographic Fragment Screening of a Bifunctional Proline Catabolic Enzyme Reveals New Inhibitor Templates for Proline Dehydrogenase and L-Glutamate-gamma-semialdehyde Dehydrogenase.
Molecules, 29, 2024
|
|
9E0C
 
 | | Structure of proline utilization A complexed with 1-benzofuran-5-ylmethanol | | Descriptor: | (1-benzofuran-5-yl)methanol, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2024-10-17 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystallographic Fragment Screening of a Bifunctional Proline Catabolic Enzyme Reveals New Inhibitor Templates for Proline Dehydrogenase and L-Glutamate-gamma-semialdehyde Dehydrogenase.
Molecules, 29, 2024
|
|
3FFI
 
 | | HIV-1 RT with pyridone non-nucleoside inhibitor | | Descriptor: | 3-chloro-5-({6-[2-(3,4-dihydroisoquinolin-2(1H)-yl)-2-oxoethyl]-3-(dimethylamino)-2-oxo-1,2-dihydropyridin-4-yl}oxy)benzonitrile, RT p51, Reverse transcriptase/ribonuclease H | | Authors: | Harris, S.F, Villasenor, A. | | Deposit date: | 2008-12-03 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pyridone Diaryl Ether Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase
To be Published
|
|
3F82
 
 | |
5IN3
 
 | | Crystal structure of glucose-1-phosphate bound nucleotidylated human galactose-1-phosphate uridylyltransferase | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-alpha-D-glucopyranose, 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Kopec, J, McCorvie, T, Tallant, C, Velupillai, S, Shrestha, L, Fitzpatrick, F, Patel, D, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular basis of classic galactosemia from the structure of human galactose 1-phosphate uridylyltransferase.
Hum.Mol.Genet., 25, 2016
|
|
7ME8
 
 | | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the beta-site, sodium ion at the metal coordination site and dual beta-Q114 rotamer conformation at 1.60 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the beta-site, sodium ion at the metal coordination site and dual beta-Q114 rotamer conformation at 1.60 Angstrom resolution.
To be Published
|
|
7WQ0
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a fragment and ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPM
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a fragment and ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPL
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPJ
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, Methionine--tRNA ligase | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
8QGA
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-methyl-amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-naphthalen-1-yl-urea, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QGE
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-pent-4-ynyl-amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-ethyl-urea, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
