6V5N
 
 | | EGFR(T790M/V948R) in complex with LN2084 | | Descriptor: | 3-[4-(4-fluorophenyl)-5-(2-phenyl-1H-pyrrolo[2,3-b]pyridin-4-yl)-1H-imidazol-2-yl]propan-1-ol, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6G6H
 
 | |
7C46
 
 | |
6MKQ
 
 | | Carbapenemase VCC-1 bound to avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Mark, B.L, Vadlamani, G. | | Deposit date: | 2018-09-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for the Potent Inhibition of the Emerging Carbapenemase VCC-1 by Avibactam.
Antimicrob. Agents Chemother., 63, 2019
|
|
5UV4
 
 | | Crystal Structure of Maize SIRK1 (sucrose-induced receptor kinase 1) kinase domain bound to AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Putative leucine-rich repeat protein kinase family protein | | Authors: | Counago, R.M, Aquino, B, Massirer, K.B, Gileadi, O, Arruda, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-17 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Maize SIRK1 (sucrose-induced receptor kinase 1) kinase domain bound to AMP-PNP
To Be Published
|
|
6ML6
 
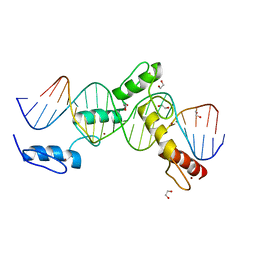 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpA 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*(5CM)P*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6G0I
 
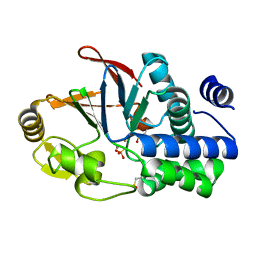 | | Active Fe-PP1 | | Descriptor: | FE (III) ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Salvi, F, Barabas, O, Koehn, M. | | Deposit date: | 2018-03-18 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of stably incorporated iron on protein phosphatase-1 structure and activity.
FEBS Lett., 592, 2018
|
|
8KBD
 
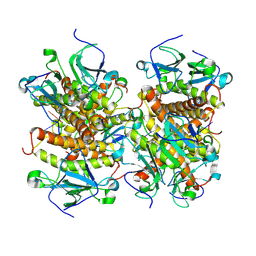 | | Structure of CmTad1 complexed with cAAG | | Descriptor: | Thoeris anti-defense 1, ZINC ION, cAAG | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Single phage proteins sequester signals from TIR and CGAS-like enzymes
Nature, 2024
|
|
6Y32
 
 | | Structure of the GTPase heterodimer of human SRP54 and SRalpha | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, SULFATE ION, ... | | Authors: | Juaire, K.D, Becker, M.M.M, Wild, K, Sinning, I. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Functional Impact of SRP54 Mutations Causing Severe Congenital Neutropenia.
Structure, 29, 2021
|
|
8KCK
 
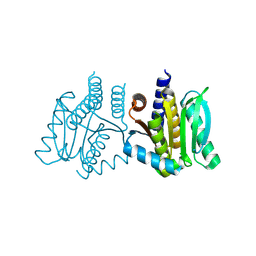 | | De novo design protein -N9 | | Descriptor: | De novo design protein -N9 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | De novo design protein -N9
To Be Published
|
|
6V80
 
 | |
6MLX
 
 | | Crystal structure of T. pallidum Leucine Rich Repeat protein (TpLRR) | | Descriptor: | Leucine-rich repeat protein TpLRR | | Authors: | Ramaswamy, R, Loveless, B.C, Houston, S, Cameron, C.E, Boulanger, M.J. | | Deposit date: | 2018-09-28 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of Treponema pallidum Tp0225 reveals an unexpected leucine-rich repeat architecture.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6PG3
 
 | |
6P82
 
 | | Structure of P. aeruginosa ATCC27853 CdnD, Apo form 1 | | Descriptor: | Nucleotidyltransferase, SULFATE ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
8KC0
 
 | | De novo design protein -NB8 | | Descriptor: | De novo design protein -NB8 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo design protein -NB8
To Be Published
|
|
5HRQ
 
 | | Insulin with proline analog HzP at position B28 in the R6 state | | Descriptor: | CHLORIDE ION, Insulin A-Chain, Insulin B-Chain, ... | | Authors: | Lieblich, S.A, Fang, K.Y, Cahn, J.K.B, Tirrell, D.A. | | Deposit date: | 2016-01-24 | | Release date: | 2017-01-25 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.
J. Am. Chem. Soc., 139, 2017
|
|
6Y33
 
 | | Streptavidin mutant S112R with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6G1L
 
 | | MITF/CLEARbox structure | | Descriptor: | CLEAR-box, Microphthalmia-associated transcription factor | | Authors: | Pogenberg, V, Wilmanns, M. | | Deposit date: | 2018-03-21 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | MITF has a central role in regulating starvation-induced autophagy in melanoma.
Sci Rep, 9, 2019
|
|
7C8L
 
 | | Hybrid designing of potent inhibitors of Striga strigolactone receptor ShHTL7 | | Descriptor: | 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, GLYCEROL, Hyposensitive to light 7, ... | | Authors: | Shahul Hameed, U.F, Arold, S.T. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Rational design of Striga hermonthica-specific seed germination inhibitors.
Plant Physiol., 188, 2022
|
|
6V95
 
 | | Peanut lectin complexed with divalent N-beta-D-galactopyranosyl-L-tartaramidoyl derivative (diNGT) | | Descriptor: | (2R,3R)-N-[(1-{(3S,3aR,6S,6aR)-6-[4-({[(2R,3R)-2,3-dihydroxy-4-oxo-4-{[(2R,3R,4R,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]amino}butanoyl]amino}methyl)-1H-1,2,3-triazol-1-yl]hexahydrofuro[3,2-b]furan-3-yl}-1H-1,2,3-triazol-4-yl)methyl]-2,3-dihydroxy-N'-[(2R,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]butanediamide (non-preferred name), CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-13 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5KUU
 
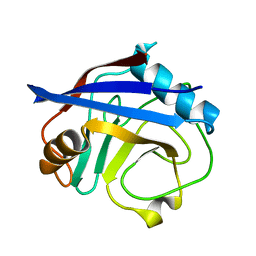 | | Human cyclophilin A at 100K, Data set 7 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5V2T
 
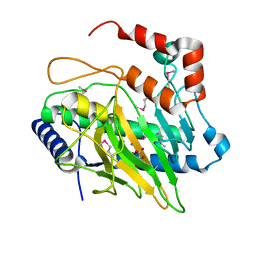 | | Ethylene forming enzyme in complex with manganese and tartrate | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, L(+)-TARTARIC ACID, MANGANESE (II) ION | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.227 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
6V7J
 
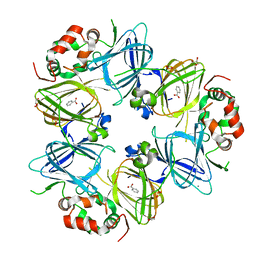 | | The C2221 crystal form of canavalin at 173 K | | Descriptor: | BENZOIC ACID, CALCIUM ION, Canavalin, ... | | Authors: | McPherson, A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of benzoic acid and anions within the cupin domains of the vicilin protein canavalin from jack bean (Canavalia ensiformis): Crystal structures.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
6Y49
 
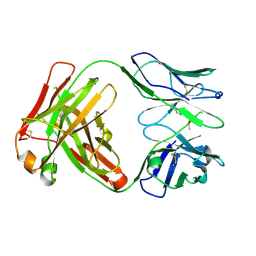 | | Crystal structure of the paraoxon-modified A.17kappa antibody FAB fragment | | Descriptor: | A.17kappa antibody FAB fragment - Heavy Chain, A.17kappa antibody FAB fragment - Light Chain, DIETHYL PHOSPHONATE | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-19 | | Release date: | 2020-09-16 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5DHK
 
 | |
