6L81
 
 | | Crystal structure of Homo sapiens GCP5 N-terminus and Mozart1 | | Descriptor: | Gamma-tubulin complex component 5, Mitotic-spindle organizing protein 1 | | Authors: | Huang, T.L, Wang, H.J, Wang, S.W, Hsia, K.C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.19651 Å) | | Cite: | Promiscuous Binding of Microprotein Mozart1 to gamma-Tubulin Complex Mediates Specific Subcellular Targeting to Control Microtubule Array Formation.
Cell Rep, 31, 2020
|
|
6O3Z
 
 | | Crystal structure of RORgt with 3-cyano-N-(3-{[(3S)-4-(cyclopentanecarbonyl)-3-methylpiperazin-1-yl]methyl}-5-fluoro-2-methylphenyl)benzamide (compound 1) | | Descriptor: | 1,2-ETHANEDIOL, 3-cyano-N-(3-{[(3S)-4-(cyclopentanecarbonyl)-3-methylpiperazin-1-yl]methyl}-5-fluoro-2-methylphenyl)benzamide, RAR-related orphan receptor C isoform a variant | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of [1,2,4]Triazolo[1,5-a]pyridine Derivatives as Potent and Orally Bioavailable ROR gamma t Inverse Agonists.
Acs Med.Chem.Lett., 11, 2020
|
|
8F5V
 
 | |
7AJR
 
 | | Virtual screening approach leading to the identification of a novel and tractable series of Pseudomonas aeruginosa elastase inhibitors | | Descriptor: | 2-[2-(1,3-benzothiazol-2-ylmethylcarbamoyl)-1,3-dihydroinden-2-yl]ethanoic acid, Keratinase KP2, SULFATE ION, ... | | Authors: | Leiris, S, Davies, D.T, Sprinsky, N, Castandet, J, Behria, L, Bodnarchuk, M.S, Sutton, J.M, Mullins, T.M.G, Jones, M.W, Forrest, A.K, Pallin, T.D, Karunakar, P, Martha, S.K, Parusharamulu, B, Ramula, R, Kotha, V, Pottabathini, N, Pothukanuri, S, Lemonnier, M, Everett, M. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Virtual Screening Approach to Identifying a Novel and Tractable Series of Pseudomonas aeruginosa Elastase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
6GIN
 
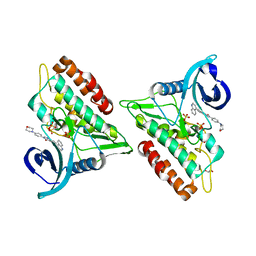 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with an Quinazolinone based ALK2 inhibitor with a 4-morpholinophenyl solvent accessible group. | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-morpholin-4-ylphenyl)-6-quinolin-4-yl-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Kopec, J, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
5B5O
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with N-phenyl-4-((4H-1,2,4-triazol-3-ylsulfanyl)methyl)-1,3-thiazol-2-amine | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Collagenase 3, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2016-05-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of Novel, Highly Potent, and Selective Matrix Metalloproteinase (MMP)-13 Inhibitors with a 1,2,4-Triazol-3-yl Moiety as a Zinc Binding Group Using a Structure-Based Design Approach
J. Med. Chem., 60, 2017
|
|
7L0L
 
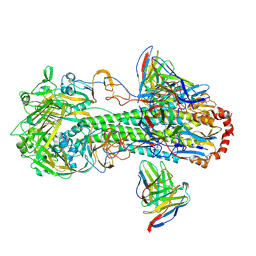 | | Cryo-EM structure of the VRC316 clinical trial, vaccine-elicited, human antibody 316-310-1B11 in complex with an H2 CAN05 HA trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 316-310-1B11 Heavy Chain, 316-310-1B11 Light Chain, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-11-03 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | A single residue in influenza virus H2 hemagglutinin enhances the breadth of the B cell response elicited by H2 vaccination.
Nat Med, 28, 2022
|
|
6P38
 
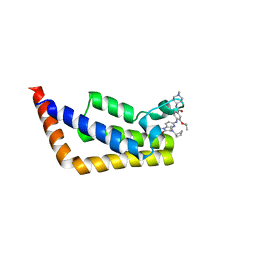 | | Crystal Structure Analysis of TAF1 Bromodomain | | Descriptor: | 4-{[(3R)-4-cyclopentyl-1,3-dimethyl-2-oxo-1,2,3,4-tetrahydropyrido[2,3-b]pyrazin-6-yl]amino}-N-(1-methylpiperidin-4-yl)-3-[(propan-2-yl)oxy]benzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual Inhibition of TAF1 and BET Bromodomains from the BI-2536 Kinase Inhibitor Scaffold.
Acs Med.Chem.Lett., 10, 2019
|
|
5YP6
 
 | | RORgamma (263-509) complexed with SRC2 and Compound 6 | | Descriptor: | N-[3'-cyano-4'-(2-methylpropyl)-2-(trifluoromethyl)biphenyl-4-yl]-2-[4-(ethylsulfonyl)phenyl]acetamide, Nuclear receptor ROR-gamma, SRC2 | | Authors: | Gao, M, Cai, W. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | From ROR gamma t Agonist to Two Types of ROR gamma t Inverse Agonists
ACS Med Chem Lett, 9, 2018
|
|
7BF5
 
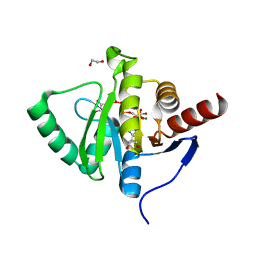 | | Crystal structure of SARS-CoV-2 macrodomain in complex with ADP-ribose-phosphate (ADP-ribose-2'-phosphate, ADPRP) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NSP3 macrodomain, ... | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
7BF3
 
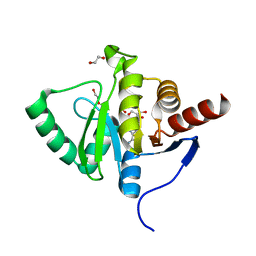 | | Crystal structure of SARS-CoV-2 macrodomain in complex with adenosine | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, MAGNESIUM ION, ... | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
7BF6
 
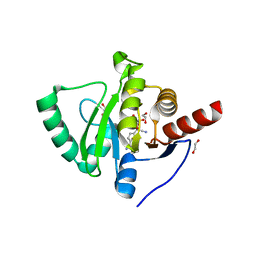 | | Crystal structure of SARS-CoV-2 macrodomain in complex with remdesivir metabolite GS-441524 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile, 1,2-ETHANEDIOL, Papain-like protease nsp3 | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6TE6
 
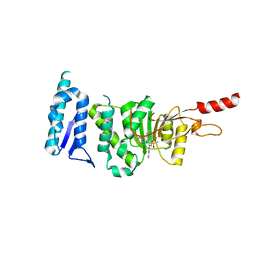 | | Crystal structure of Dot1L in complex with an inhibitor (compound 3). | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, ~{N}1-[(~{S})-(3-chlorophenyl)-pyridin-2-yl-methyl]-4-methylsulfonyl-~{N}2-pyrimidin-2-yl-benzene-1,2-diamine | | Authors: | Scheufler, C, Stauffer, F, Be, C, Moebitz, H. | | Deposit date: | 2019-11-11 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | New Potent DOT1L Inhibitors forin VivoEvaluation in Mouse.
Acs Med.Chem.Lett., 10, 2019
|
|
7BF4
 
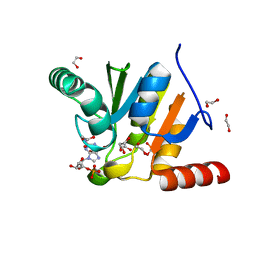 | | Crystal structure of SARS-CoV-2 macrodomain in complex with GMP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, NSP3 macrodomain | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
5C0R
 
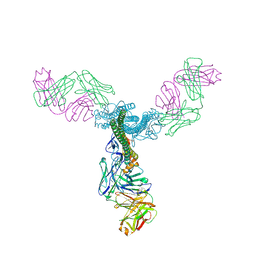 | | Crystal Structure of a Generation 3 Influenza Hemagglutinin Stabilized Stem Complexed with the Broadly Neutralizing Antibody C179 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C179 Fab heavy chain, ... | | Authors: | Boyington, J.C, kwong, P.D, Nabel, G.J, Mascola, J.R. | | Deposit date: | 2015-06-12 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection.
Nat. Med., 21, 2015
|
|
8ATM
 
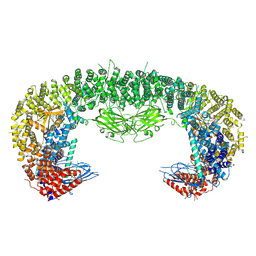 | |
8ATO
 
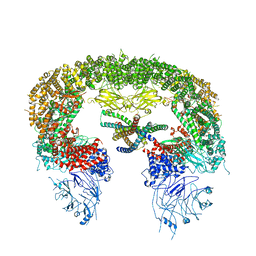 | |
5YP5
 
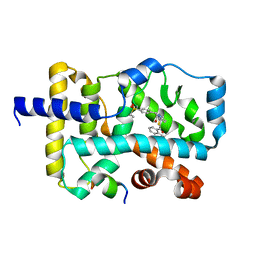 | | Crystal structure of RORgamma complexed with SRC2 and compound 5d | | Descriptor: | 2-[4-(ethylsulfonyl)phenyl]-N-{5-[2-(2-methylpropyl)benzoyl]-4-phenyl-1,3-thiazol-2-yl}acetamide, Nuclear receptor ROR-gamma, SRC2-2 peptide | | Authors: | Gao, M, Cai, W, Chunwa, C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | From ROR gamma t Agonist to Two Types of ROR gamma t Inverse Agonists
ACS Med Chem Lett, 9, 2018
|
|
8FX9
 
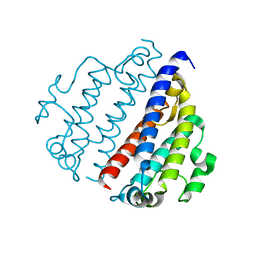 | |
6CAD
 
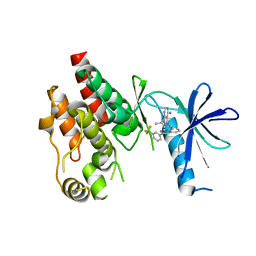 | | Crystal structure of RAF kinase domain bound to the inhibitor 2a | | Descriptor: | 1-(propan-2-yl)-3-({3-[3-(trifluoromethyl)phenyl]isoquinolin-8-yl}ethynyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Kurinov, I, Assadieskandar, A, Yu, C, Liu, X, Chen, Y.-C, Prakash, G.K.S, Zhang, C, SIcheri, F. | | Deposit date: | 2018-01-30 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Effects of rigidity on the selectivity of protein kinase inhibitors.
Eur J Med Chem, 146, 2018
|
|
8ECM
 
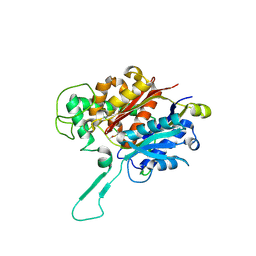 | |
5M5J
 
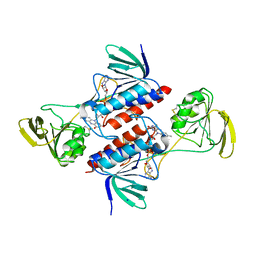 | | Thioredoxin reductase from Giardia duodenalis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SODIUM ION, Thioredoxin reductase | | Authors: | Fiorillo, A, Ilari, A, Lalle, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural characterization of Giardia duodenalis thioredoxin reductase (gTrxR) and computational analysis of its interaction with NBDHEX.
Eur J Med Chem, 135, 2017
|
|
7GB5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-0da5ad92-2 (Mpro-x10201) | | Descriptor: | 2-(3-chlorophenyl)-N-(pyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.261 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBL
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with RAL-MED-2de63afb-1 (Mpro-x10371) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(3-{[(2S)-4-oxoazetidin-2-yl]oxy}phenyl)-2-(pyridin-3-yl)acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GDK
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-0da5ad92-21 (Mpro-x11025) | | Descriptor: | 2-(3-chlorophenyl)-N-(3-methyl-1H-pyrazol-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
