6ZJ9
 
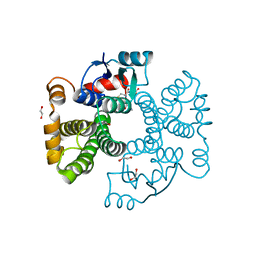 | | Crystal structure of Equus ferus caballus glutathione transferase A3-3 in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase | | Authors: | Skerlova, J, Ismail, A, Lindstrom, H, Sjodin, B, Mannervik, B, Stenmark, P. | | Deposit date: | 2020-06-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of the inhibition of equine glutathione transferase A3-3 by organotin endocrine disrupting pollutants.
Environ Pollut, 268, 2021
|
|
6Z8X
 
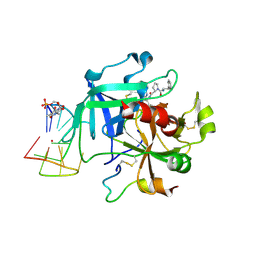 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3Leu), which contains leucyl amide in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
6ZL6
 
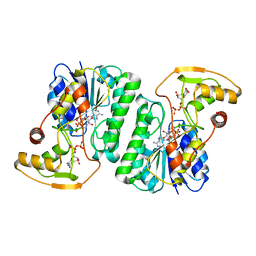 | |
8G6A
 
 | | Wildtype PTP1b in complex with DES6016 | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Greisman, J.B, Willmore, L, Yeh, C.Y, Giordanetto, F, Shahamadtar, S, Nisonoff, H, Maragakis, P, Shaw, D.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography.
J.Chem.Inf.Model., 63, 2023
|
|
8G3V
 
 | |
6XV6
 
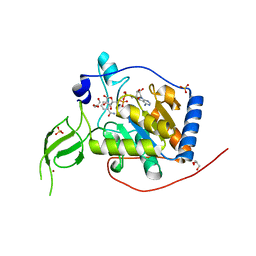 | | Human Sirt6 3-318 in complex with ADP-ribose | | Descriptor: | GLYCEROL, NAD-dependent protein deacetylase sirtuin-6, NICOTINAMIDE, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2020-01-21 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Binding site for activator MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
8G3Z
 
 | |
8GXD
 
 | | L-LEUCINE DEHYDROGENASE FROM EXIGUOBACTERIUM SIBIRICUM | | Descriptor: | CALCIUM ION, GLYCEROL, Glu/Leu/Phe/Val dehydrogenase | | Authors: | Mu, X, Nie, Y, Wu, T, Wang, Y, Zhang, N, Yin, D, Xu, Y. | | Deposit date: | 2022-09-19 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Reshaping Substrate-Binding Pocket of Leucine Dehydrogenase for Bidirectionally Accessing Structurally Diverse Substrates
Acs Catalysis, 13, 2023
|
|
8GP6
 
 | |
6XXT
 
 | |
1HR7
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant | | Descriptor: | MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ZINC ION | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
8G2G
 
 | | Crystal structure of PRMT3 with compound YD1113 | | Descriptor: | 5'-S-[2-(benzylcarbamamido)ethyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3, SULFATE ION | | Authors: | Song, X, Dong, A, Arrowsmith, C.H, Edwards, A.M, Deng, Y, Huang, R, Min, J. | | Deposit date: | 2023-02-03 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
8G3T
 
 | | MBP-Mcl1 in complex with ligand 12 | | Descriptor: | (1'S,3aS,5R,16R,17S,19Z,21R,21aR)-6'-chloro-20-fluoro-21-methoxy-16,17-dimethyl-2,3,3',3a,4',16,17,18,21,21a-decahydro-2'H,6H,8H-15lambda~6~-spiro[10,12-etheno-15lambda~6~-furo[3,2-i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-7,1'-naphthalene]-13,15,15(4H,14H)-trione, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G68
 
 | | Wildtype PTP1b in complex with DES5742 | | Descriptor: | 4-(3-ethyl-5-methyl-1H-pyrazol-1-yl)aniline, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Greisman, J.B, Willmore, L, Yeh, C.Y, Giordanetto, F, Shahamadtar, S, Nisonoff, H, Maragakis, P, Shaw, D.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography.
J.Chem.Inf.Model., 63, 2023
|
|
8GMN
 
 | | Crystal structure of human C1s in complex with inhibitor | | Descriptor: | Complement C1s subcomponent, SULFATE ION, [4-(1-aminophthalazin-6-yl)piperazin-1-yl](2-methylphenyl)methanone | | Authors: | Dougan, D.R, Lane, W. | | Deposit date: | 2023-03-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Novel Series of Potent, Selective, Orally Available, and Brain-Penetrable C1s Inhibitors for Modulation of the Complement Pathway.
J.Med.Chem., 66, 2023
|
|
8G65
 
 | | Wildtype PTP1b in complex with DES4799 | | Descriptor: | 4-(3,5-dimethyl-1H-pyrazol-1-yl)aniline, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Greisman, J.B, Willmore, L, Yeh, C.Y, Giordanetto, F, Shahamadtar, S, Nisonoff, H, Maragakis, P, Shaw, D.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography.
J.Chem.Inf.Model., 63, 2023
|
|
8G45
 
 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with SGC-UBD253 chemical probe | | Descriptor: | 3-[8-chloro-3-(2-{[(2-methoxyphenyl)methyl]amino}-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl]propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
8G3Y
 
 | | MBP-Mcl1 in complex with ligand 34 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G3S
 
 | | MBP-Mcl1 in complex with ligand 11 | | Descriptor: | (1'S,3aS,5R,16R,17S,19E,21S,21aR)-6'-chloro-21-methoxy-16,17-dimethyl-2,3,3',3a,4',16,17,18,21,21a-decahydro-2'H,6H,8H-15lambda~6~-spiro[10,12-etheno-15lambda~6~-furo[3,2-i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-7,1'-naphthalene]-13,15,15(4H,14H)-trione, FORMIC ACID, Maltodextrin-binding protein, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
1HI6
 
 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE | | Descriptor: | IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN), IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN), PEPTIDE 5 | | Authors: | Hahn, M, Wessner, H, Schneider-Mergener, J, Hohne, W. | | Deposit date: | 2001-01-02 | | Release date: | 2001-02-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic Analysis of Anti-P24 (HIV-1) Monoclonal Antibody Cross-Reactivity and Polyspecificity
Cell(Cambridge,Mass.), 91, 1997
|
|
8G3U
 
 | | MBP-Mcl1 in complex with ligand 21 | | Descriptor: | (1'S,3aS,5R,16R,17S,19Z,21R,21aR)-6'-chloro-20-fluoro-21-{[(5S,9aS)-hexahydropyrazino[2,1-c][1,4]oxazin-8(1H)-yl]methyl}-21-methoxy-16,17-dimethyl-2,3,3',3a,4',16,17,18,21,21a-decahydro-2'H,6H,8H-15lambda~6~-spiro[10,12-(ethanediylidene)-15lambda~6~-furo[3,2-i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-7,1'-naphthalene]-13,15,15(4H,14H)-trione, 1,2-ETHANEDIOL, Maltodextrin-binding protein, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G3X
 
 | | MBP-Mcl1 in complex with ligand 32 | | Descriptor: | 1,2-ETHANEDIOL, Maltodextrin-binding protein, Induced myeloid leukemia cell differentiation protein Mcl-1 chimera, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G43
 
 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(methylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(methylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
6XPB
 
 | |
6XFJ
 
 | | Crystal structure of the type III secretion pilotin InvH | | Descriptor: | CADMIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
