8SRY
 
 | | Crystal structure of BAK-BAX heterodimer with C12E8 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, ... | | Authors: | Brouwer, J.M, Czabotar, P.E, Colman, P.M, Miller, M.S. | | Deposit date: | 2023-05-08 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
8VM2
 
 | | Crystal Structure of NRAS Q61K bound to GTP | | Descriptor: | GTPase NRas, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gebregiworgis, T, Chan, J.Y.L, Kuntz, D.A, Prive, G.G, Marshall, C.B, Ikura, M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of NRAS Q61K with a ligand-induced pocket near switch II.
Eur J Cell Biol, 103, 2024
|
|
8VQR
 
 | | Crystal structure of chimeric SARS-CoV-2 RBD complexed with chimeric raccoon dog ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsueh, F.-C, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2024-01-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.565 Å) | | Cite: | Structural basis for raccoon dog receptor recognition by SARS-CoV-2
To Be Published
|
|
6NKP
 
 | |
5E8W
 
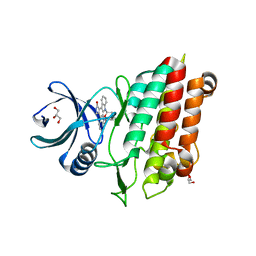 | |
7N2W
 
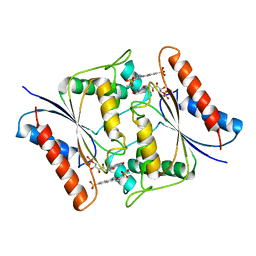 | | The crystal structure of an FMN-dependent NADH-azoreductase, AzoA in complex with Red 40 | | Descriptor: | 6-hydroxy-5-[(E)-(2-methoxy-5-methyl-4-sulfophenyl)diazenyl]naphthalene-2-sulfonic acid, FLAVIN MONONUCLEOTIDE, FMN dependent NADH:quinone oxidoreductase | | Authors: | Arcinas, A.J, Fedorov, E, Kelly, L, Almo, S.C, Ghosh, A. | | Deposit date: | 2021-05-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Uncovering a novel mechanism of enzyme activation in multimeric azoreductases
To Be Published
|
|
5VGT
 
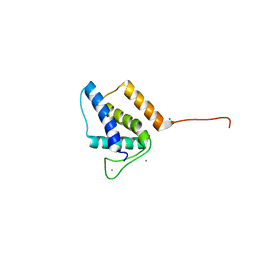 | | X-ray structure of bacteriophage Sf6 tail adaptor protein gp7 | | Descriptor: | CALCIUM ION, Gene 7 protein, MAGNESIUM ION | | Authors: | Tang, L, Liang, L, Zhao, H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | High-resolution structure of podovirus tail adaptor suggests repositioning of an octad motif that mediates the sequential tail assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5HL0
 
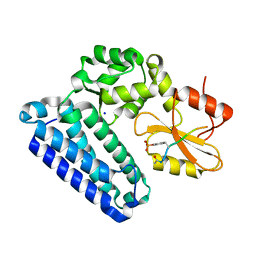 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION, Sprouty 2 (SPRY2) | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution
To Be Published
|
|
7PZ1
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH8535 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-bromanyl-2-oxidanylidene-3~{H}-benzimidazol-1-yl)-~{N}-(3-methoxy-4-methyl-phenyl)piperidine-1-carboxamide, GLYCEROL, ... | | Authors: | Scaletti, E.R, Helleday, T, Stenmark, P. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Optimization of N-Piperidinyl-Benzimidazolone Derivatives as Potent and Selective Inhibitors of 8-Oxo-Guanine DNA Glycosylase 1.
Chemmedchem, 18, 2023
|
|
8VBV
 
 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Cephalexin) inhibited form | | Descriptor: | (2S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
6NMM
 
 | |
8PX9
 
 | | Structure of the antibacterial peptide ABC transporter McjD, solved at wavelength 2.75 A | | Descriptor: | MAGNESIUM ION, Microcin-J25 export ATP-binding/permease protein McjD, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Bountra, K, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
7Q10
 
 | | Acetyltrasferase(3) type IIIa in complex with 3-N-methyl-nemycin B | | Descriptor: | 3N methyl nemycin B, ACETATE ION, Aminoglycoside N(3)-acetyltransferase III, ... | | Authors: | Pontillo, N, Guskov, A. | | Deposit date: | 2021-10-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 3-N-alkylation in aminoglycoside antibiotic neomycin B overcomes bacterial resistance mediated by acetyltransferase (3) IIIa
To Be Published
|
|
5VGY
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5S)-5-[(2,4-dimethoxyphenyl)methyl]-5-hydroxy-2-sulfanylideneimidazolidin-4-one, Carbonic anhydrase 2, SODIUM ION, ... | | Authors: | Peat, T.S. | | Deposit date: | 2017-04-11 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
6NN1
 
 | | Xanthomonas citri PGM Apo-Dephospho | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stiers, K.M, Beamer, L.J. | | Deposit date: | 2019-01-14 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and dynamical description of the enzymatic reaction of a phosphohexomutase.
Struct Dyn., 6, 2019
|
|
7UAJ
 
 | | Crystal structure of apo HPV16 E6 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein E6, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Shen, Q, Leonard, P.G, Cross, J.B. | | Deposit date: | 2022-03-13 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Disorder-to-order transition of the interdomain linker of HPV E6 upon E6AP binding reshapes p53 binding pocket
To Be Published
|
|
5HW5
 
 | |
7YRE
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, staygold(N137A,Q140S,Y187F) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
7UW2
 
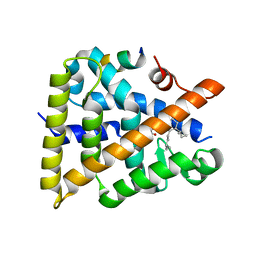 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with UAB116 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-8-{3-[(2S)-butan-2-yl]-2-(3-methylbutyl)cyclohex-2-en-1-ylidene}-3,7-dimethylocta-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chattopadhyay, D, Yang, Z, Atigadda, V. | | Deposit date: | 2022-05-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Conformationally Defined Rexinoids for the Prevention of Inflammation and Nonmelanoma Skin Cancers.
J.Med.Chem., 65, 2022
|
|
5EAC
 
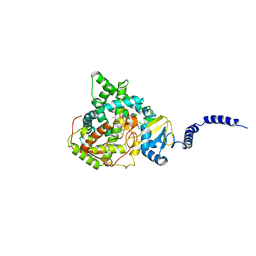 | | Saccharomyces cerevisiae CYP51 complexed with the plant pathogen inhibitor R-tebuconazole | | Descriptor: | (3~{R})-1-(4-chlorophenyl)-4,4-dimethyl-3-(1,2,4-triazol-1-ylmethyl)pentan-3-ol, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tyndall, J.D.A, Sabherwal, M, Sagatova, A.A, Keniya, M.V, Wilson, R.K, Woods, M.V, Monk, B.C. | | Deposit date: | 2015-10-16 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural and Functional Elucidation of Yeast Lanosterol 14 alpha-Demethylase in Complex with Agrochemical Antifungals.
PLoS ONE, 11, 2016
|
|
5HLD
 
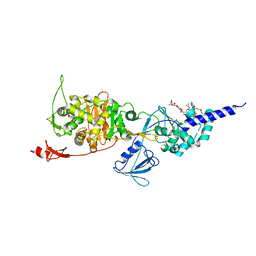 | | E. coli PBP1b in complex with acyl-CENTA and moenomycin | | Descriptor: | (2S)-5-methylidene-2-{(1R)-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2016-01-14 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Escherichia coli Penicillin-Binding Protein 1B: Structural Insights into Inhibition.
J. Biol. Chem., 2016
|
|
8PXC
 
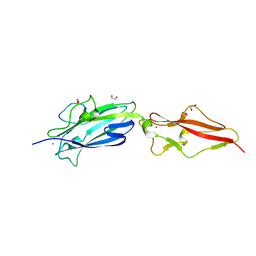 | | Structure of Fap1, a domain of the accessory Sec-dependent serine-rich glycoprotein adhesin from Streptococcus oralis, solved at wavelength 3.06 A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fap1 | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Owen, C.D, Walsh, M.A, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
7PI6
 
 | | Trypanosoma brucei ISG65 bound to human complement C3d | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 65 kDa invariant surface glycoprotein, Complement C3dg fragment, ... | | Authors: | Cook, A.D, Higgins, M.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Invariant surface glycoprotein 65 of Trypanosoma brucei is a complement C3 receptor.
Nat Commun, 13, 2022
|
|
8PNY
 
 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens complexed with phenylhydrazine and in its apo form | | Descriptor: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
7U9K
 
 | | Staphylococcus aureus D-alanine-D-alanine ligase in complex with ATP, D-ala-D-ala, Mg2+ and K+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, D-alanine--D-alanine ligase, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided design and synthesis of ATP-competitive N-acyl-substituted sulfamide d-alanine-d-alanine ligase inhibitors.
Bioorg.Med.Chem., 96, 2023
|
|
