1UE9
 
 | | Solution structure of the fourth SH3 domain of human intersectin 2 (KIAA1256) | | Descriptor: | Intersectin 2 | | Authors: | Tochio, N, Kigawa, T, Koshiba, S, Kobayashi, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-09 | | Release date: | 2003-11-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fourth SH3 domain of human intersectin 2 (KIAA1256)
To be Published
|
|
8B8R
 
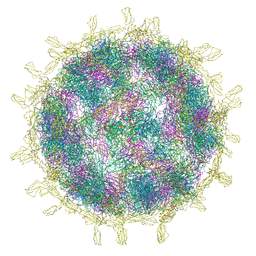 | | Complex of Echovirus 11 with its attaching receptor decay-accelerating factor (CD55) | | Descriptor: | DECAY ACCELERATING FACTOR (CD55), SPHINGOSINE, VP1, ... | | Authors: | Stuart, D.I, Ren, J, Zhou, D, Qin, L. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Viruses, 14, 2022
|
|
8AV6
 
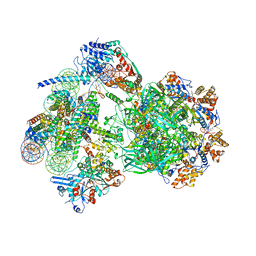 | | CryoEM structure of INO80 core nucleosome complex in closed grappler conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DASH complex subunit DAD4, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
1UEZ
 
 | | Solution structure of the first PDZ domain of human KIAA1526 protein | | Descriptor: | KIAA1526 Protein | | Authors: | Li, H, Kigawa, T, Muto, Y, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-22 | | Release date: | 2003-11-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first PDZ domain of human KIAA1526 protein
To be Published
|
|
1UFL
 
 | | Crystal Structure of TT1020 from Thermus thermophilus HB8 | | Descriptor: | Nitrogen regulatory protein P-II | | Authors: | Wang, H, Sakai, H, Hori-Takemoto, C, Kaminishi, T, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-31 | | Release date: | 2003-11-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the signal transducing protein GlnK from Thermus thermophilus HB8.
J.Struct.Biol., 149, 2005
|
|
8B6L
 
 | | Subtomogram average of the human Sec61-TRAP-OSTA-translocon | | Descriptor: | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit, Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1, Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2, ... | | Authors: | Gemmer, M, Fedry, J.M.M, Forster, F.G. | | Deposit date: | 2022-09-27 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Visualization of translation and protein biogenesis at the ER membrane.
Nature, 614, 2023
|
|
4DFA
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A/L36A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Clark, I.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
8AYY
 
 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and Pleconaril | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
1UFX
 
 | | Solution structure of the third PDZ domain of human KIAA1526 protein | | Descriptor: | KIAA1526 protein | | Authors: | Tochio, N, Kobayashi, N, Koshiba, S, Kigawa, T, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third PDZ domain of human KIAA1526 protein
To be Published
|
|
1U6Q
 
 | | Substituted 2-Naphthamadine inhibitors of Urokinase | | Descriptor: | TRANS-6-(2-PHENYLCYCLOPROPYL)-NAPHTHALENE-2-CARBOXAMIDINE, Urokinase-type plasminogen activator | | Authors: | Bruncko, M, McClellan, W, Wendt, M.D, Sauer, D.R, Geyer, A, Dalton, C.R, Kaminski, M.K, Nienaber, V.L, Rockway, T.R, Giranda, V.L. | | Deposit date: | 2004-07-30 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Naphthamidine urokinase plasminogen activator inhibitors with improved pharmacokinetic properties.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
8AYX
 
 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
1U72
 
 | |
8BFP
 
 | | Jumbo Phage phi-kp24 empty capsid pentamer hexamers | | Descriptor: | Major head protein | | Authors: | Ouyang, R. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers.
Nat Commun, 13, 2022
|
|
8B2N
 
 | | Potempin A (PotA) from Tannerella forsythia in complex with the catalytic domain of human MMP-12 | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, Tannerella forsythia potempin A (PotA), ... | | Authors: | Potempa, J, Ksiazek, M, Goulas, T, Cuppari, A, Rodriguez-Banqueri, A, Arolas, J.L, Lopez-Pelegrin, M, Garcia-Ferrer, I, Guevara, T. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A unique network of attack, defence and competence on the outer membrane of the periodontitis pathogen Tannerella forsythia.
Chem Sci, 14, 2023
|
|
1U78
 
 | |
3SKD
 
 | |
1U7F
 
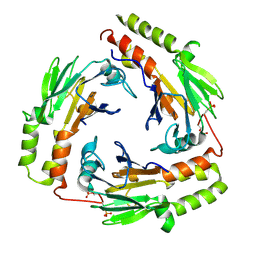 | | Crystal Structure of the phosphorylated Smad3/Smad4 heterotrimeric complex | | Descriptor: | Mothers against decapentaplegic homolog 3, Mothers against decapentaplegic homolog 4 | | Authors: | Chacko, B.M, Qin, B.Y, Tiwari, A, Shi, G, Lam, S, Hayward, L.J, de Caestecker, M, Lin, K. | | Deposit date: | 2004-08-03 | | Release date: | 2004-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of heteromeric smad protein assembly in tgf-Beta signaling
Mol.Cell, 15, 2004
|
|
3SJV
 
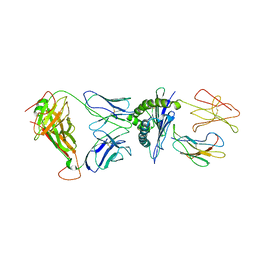 | | Crystal structure of the RL42 TCR in complex with HLA-B8-FLR | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 3, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
1U7R
 
 | |
2IHP
 
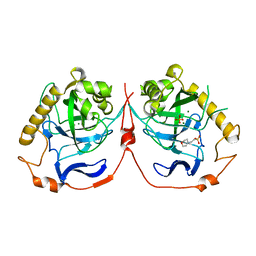 | | Yeast inorganic pyrophosphatase with magnesium and phosphate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Inorganic pyrophosphatase, MAGNESIUM ION, ... | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-09-27 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
4DGZ
 
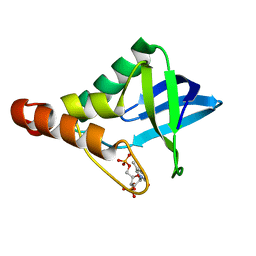 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A/L125A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Nam, S.P, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
8ASV
 
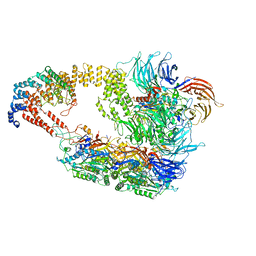 | | Cryo-EM structure of yeast Elongator complex | | Descriptor: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-21 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
1U7X
 
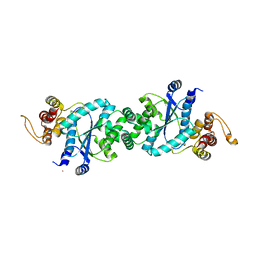 | | crystal structure of a mutant M. jannashii tyrosyl-tRNA synthetase specific for O-methyl-tyrosine | | Descriptor: | POTASSIUM ION, Tyrosyl-tRNA synthetase | | Authors: | Zhang, Y, Wang, L, Schultz, P.G, Wilson, I.A. | | Deposit date: | 2004-08-04 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of apo wild-type M. jannaschii tyrosyl-tRNA synthetase (TyrRS) and an engineered TyrRS specific for O-methyl-L-tyrosine.
Protein Sci., 14, 2005
|
|
8BBH
 
 | |
1UGL
 
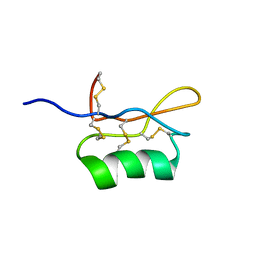 | | Solution structure of S8-SP11 | | Descriptor: | S-locus pollen protein | | Authors: | Mishima, M, Takayama, S, Sasaki, K, Jee, J.G, Kojima, C, Isogai, A, Shirakawa, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-16 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the Male Determinant Factor for Brassica Self-incompatibility
J.Biol.Chem., 278, 2003
|
|
