1OWM
 
 | | DATA1:DNA photolyase / received X-rays dose 1.2 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWN
 
 | | DATA3:DNA photolyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWO
 
 | | DATA4:photoreduced DNA photolyase / received X-rays dose 1.2 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PNQ
 
 | | Crystal structure of R. rubrum transhydrogenase domain III bound to NADPH | | Descriptor: | NAD(P) transhydrogenase subunit beta, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaresan, V, Yamaguchi, M, Chartron, J, Stout, C.D. | | Deposit date: | 2003-06-12 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Change in the NADP(H) Binding Domain of Transhydrogenase Defines Four States
Biochemistry, 42, 2003
|
|
1PPK
 
 | |
1PPI
 
 | | THE ACTIVE CENTER OF A MAMMALIAN ALPHA-AMYLASE. THE STRUCTURE OF THE COMPLEX OF A PANCREATIC ALPHA-AMYLASE WITH A CARBOHYDRATE INHIBITOR REFINED TO 2.2 ANGSTROMS RESOLUTION | | Descriptor: | 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose, ALPHA-AMYLASE, CALCIUM ION, ... | | Authors: | Qian, M, Haser, R, Payan, F. | | Deposit date: | 1994-02-22 | | Release date: | 1995-05-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The active center of a mammalian alpha-amylase. Structure of the complex of a pancreatic alpha-amylase with a carbohydrate inhibitor refined to 2.2-A resolution.
Biochemistry, 33, 1994
|
|
1OWL
 
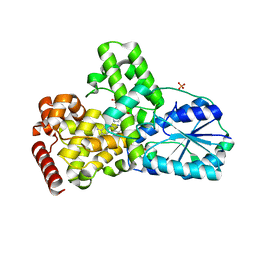 | | Structure of apophotolyase from Anacystis nidulans | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWP
 
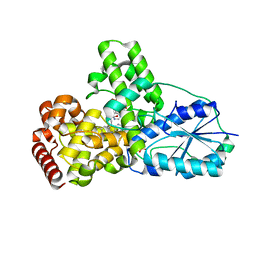 | | DATA6:photoreduced DNA pholyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PTK
 
 | |
1PPM
 
 | |
1NTC
 
 | |
1PNO
 
 | | Crystal structure of R. rubrum transhydrogenase domain III bound to NADP | | Descriptor: | NAD(P) transhydrogenase subunit beta, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaresan, V, Yamaguchi, M, Chartron, J, Stout, C.D. | | Deposit date: | 2003-06-12 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Change in the NADP(H) Binding Domain of Transhydrogenase Defines Four States
Biochemistry, 42, 2003
|
|
1PPL
 
 | |
1QU4
 
 | | CRYSTAL STRUCTURE OF TRYPANOSOMA BRUCEI ORNITHINE DECARBOXYLASE | | Descriptor: | ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Grishin, N.V, Osterman, A.L, Brooks, H.B, Phillips, M.A, Goldsmith, E.J. | | Deposit date: | 1999-07-06 | | Release date: | 1999-11-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with alpha-difluoromethylornithine.
Biochemistry, 38, 1999
|
|
4ZZL
 
 | | MexR R21W derepressor mutant causing multidrug resistance in P. aeruginosa by mexAB-oprM efflux pump expression | | Descriptor: | GLYCEROL, MULTIDRUG RESISTANCE OPERON REPRESSOR | | Authors: | Anandapadamanaban, M, Pilstal, R, Ziauddin, J.M.E, Moche, M, Wallner, B, Sunnerhagen, M. | | Deposit date: | 2015-04-10 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Mutation-Induced Population Shift in the Mexr Conformational Ensemble Disengages DNA Binding: A Novel Mechanism for Marr Family Derepression.
Structure, 24, 2016
|
|
4PEP
 
 | |
2RMP
 
 | | RMP-pepstatin A complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MUCOROPEPSIN, PEPSTATIN, ... | | Authors: | Yang, J, Quail, J.W. | | Deposit date: | 1997-05-30 | | Release date: | 1997-09-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Rhizomucor miehei aspartic proteinase complexed with the inhibitor pepstatin A at 2.7 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5A97
 
 | | Hazara virus nucleocapsid protain | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Surtees, R, Ariza, A, Hewson, R, Barr, J.N, Edwards, T.A. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of the Hazara Virus Nucleocapsid Protein.
Bmc Struct.Biol., 15, 2015
|
|
7DA5
 
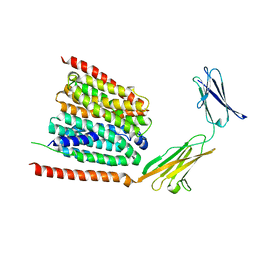 | | Cryo-EM structure of the human MCT1 D309N mutant in complex with Basigin-2 in the inward-open conformation. | | Descriptor: | Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
3VST
 
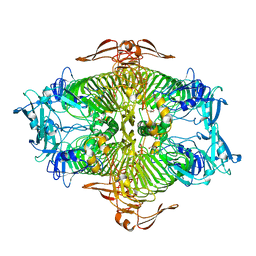 | | The complex structure of XylC with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Xylosidase | | Authors: | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | Deposit date: | 2012-05-09 | | Release date: | 2013-02-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
3VCB
 
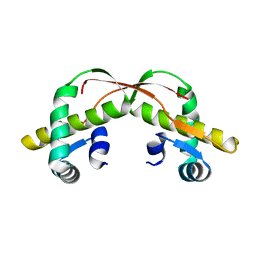 | | C425S mutant of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
3VSU
 
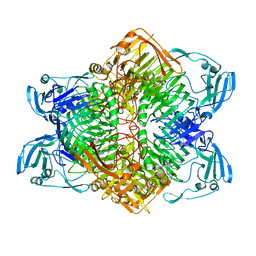 | | The complex structure of XylC with xylobiose | | Descriptor: | Xylosidase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | Deposit date: | 2012-05-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
7D7L
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in complex with YM155 | | Descriptor: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, CAFFEINE, GLYCEROL, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
1N13
 
 | | The Crystal Structure of Pyruvoyl-dependent Arginine Decarboxylase from Methanococcus jannashii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, AGMATINE, Pyruvoyl-dependent arginine decarboxylase alpha chain, ... | | Authors: | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | Deposit date: | 2002-10-16 | | Release date: | 2003-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
