2NZU
 
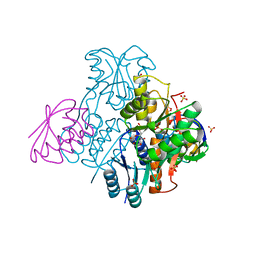 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors G6P and FBP | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, Catabolite control protein, Phosphocarrier protein HPr, ... | | Authors: | Schumacher, M.A, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-11-25 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
4AQ2
 
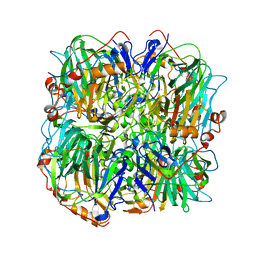 | | resting state of homogentisate 1,2-dioxygenase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, HOMOGENTISATE 1,2-DIOXYGENASE | | Authors: | Jeoung, J.-H, Lin, T.-Y, Bommer, M, Dobbek, H. | | Deposit date: | 2012-04-12 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing the Substrate-, Superoxo-, Alkylperoxo-, and Product-Bound States at the Nonheme Fe(II) Site of Homogentisate Dioxygenase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AQ6
 
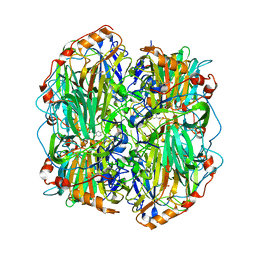 | | substrate bound homogentisate 1,2-dioxygenase | | Descriptor: | 2-(3,6-DIHYDROXYPHENYL)ACETIC ACID, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, ... | | Authors: | Jeoung, J.-H, Lin, T.-Y, Bommer, M, Dobbek, H. | | Deposit date: | 2012-04-12 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Visualizing the Substrate-, Superoxo-, Alkylperoxo- and Product-Bound States at the Non-Heme Fe(II) Site of Homogentisate Dioxygenase
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3HRY
 
 | |
5H58
 
 | | Structural and dynamics studies of the TetR family protein, CprB from Streptomyces coelicolor in complex with its biological operator sequence | | Descriptor: | CprB, DNA (5'-D(*AP*GP*GP*C*AP*GP*GP*CP*GP*GP*CP*AP*CP*GP*GP*TP*CP*TP*GP*TP*TP*GP*AP*GP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*A*CP*TP*CP*AP*AP*CP*AP*GP*AP*CP*CP*GP*TP*GP*CP*CP*GP*CP*CP*TP*GP*CP*CP*T)-3') | | Authors: | Bhukya, H, Jana, A.K, Sengupta, N, Anand, R. | | Deposit date: | 2016-11-04 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (3.991 Å) | | Cite: | Structural and dynamics studies of the TetR family protein, CprB from Streptomyces coelicolor in complex with its biological operator sequence
J. Struct. Biol., 198, 2017
|
|
2NZV
 
 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors G6P and FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Catabolite control protein, Phosphocarrier protein HPr, ... | | Authors: | Schumacher, M.A, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-11-25 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
3F8L
 
 | | Crystal Structure of the Effector Domain of PhnF from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, HTH-type transcriptional repressor phnF, SULFATE ION | | Authors: | Busby, J.N, Gebhard, S, Cook, G.M, Baker, E.N, Lott, S.J, Money, V.A. | | Deposit date: | 2008-11-12 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of PhnF, a GntR-family transcription regulator in Mycobacterium smegmatis
To be Published
|
|
3FBR
 
 | | structure of HipA-amppnp-peptide | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Serine/threonine-protein kinase toxin HipA, peptide of EF-Tu | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-11-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
7VJQ
 
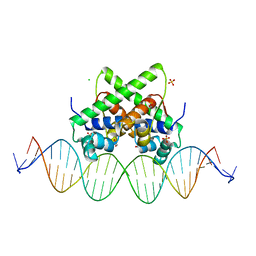 | | Pectobacterium phage ZF40 apo-aca2 complexed with 26bp DNA substrate | | Descriptor: | CHLORIDE ION, DNA (27-MER), GLYCEROL, ... | | Authors: | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
7VJO
 
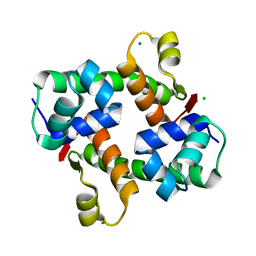 | | Pectobacterium phage ZF40 apo-Aca2 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, anti-CRISPR-associated protein Aca2 | | Authors: | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
7VJN
 
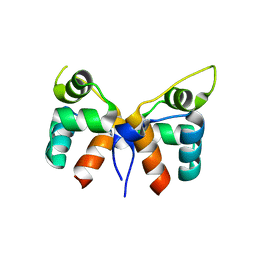 | |
7VJM
 
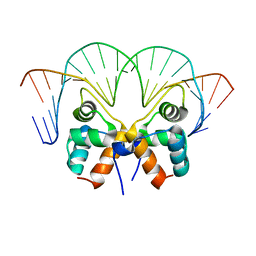 | | Aca1 in complex with 19bp palindromic DNA substrate | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*CP*AP*CP*AP*TP*TP*GP*TP*GP*CP*CP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*GP*GP*CP*AP*CP*AP*AP*TP*GP*TP*GP*CP*CP*TP*AP*A)-3'), anti-CRISPR-associated protein Aca1 | | Authors: | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
1I2A
 
 | |
1QNL
 
 | |
4YO2
 
 | | Structure of E2F8, an atypical member of E2F family of transcription factors | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*CP*CP*CP*GP*CP*CP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*GP*GP*CP*GP*GP*GP*AP*AP*AP*A)-3'), Transcription factor E2F8 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Dave, K, Schmierer, B, Popov, A, Eremina, N, Nilsson, L, Taipale, J. | | Deposit date: | 2015-03-11 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.073 Å) | | Cite: | Structural insights into the DNA-binding specificity of E2F family transcription factors.
Nat Commun, 6, 2015
|
|
8YM7
 
 | | Crystal structure of Lysine Specific Demethylase 1 (LSD1) with JH-45 | | Descriptor: | 4-[5-(4-azanylpiperidin-1-yl)-8-(4-methylphenyl)pyrido[3,4-b]pyrazin-7-yl]-2-fluoranyl-benzenecarbonitrile, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, ... | | Authors: | Zhiyan, D, Danyan, C, Hong, J, Tongchao, L, Bing, X. | | Deposit date: | 2024-03-08 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of Novel LSD1 Inhibitors for the Treatment of Autosomal Dominant Polycystic Kidney Disease
To Be Published
|
|
9FWG
 
 | | LSD1/CoREST bound to bomedemstat | | Descriptor: | Bomedemstat FAD adduct, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Speranzini, V, Mattevi, A. | | Deposit date: | 2024-06-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterization of structural, biochemical, pharmacokinetic, and pharmacodynamic properties of the LSD1 inhibitor bomedemstat in preclinical models.
Prostate, 84, 2024
|
|
1A4X
 
 | | PYRR, THE BACILLUS SUBTILIS PYRIMIDINE BIOSYNTHETIC OPERON REPRESSOR, HEXAMERIC FORM | | Descriptor: | PYRIMIDINE OPERON REGULATORY PROTEIN PYRR, SULFATE ION | | Authors: | Tomchick, D.R, Turner, R.J, Switzer, R.W, Smith, J.L. | | Deposit date: | 1998-02-08 | | Release date: | 1998-08-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase.
Structure, 6, 1998
|
|
8FY2
 
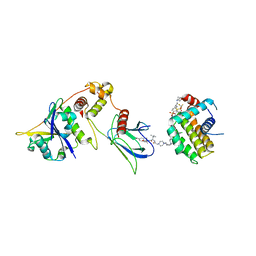 | | E3:PROTAC:target ternary complex structure (VCB/WH244/BCL-2) | | Descriptor: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | Authors: | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Ruben, E, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
8FY0
 
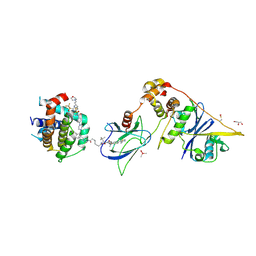 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-xL) | | Descriptor: | Bcl-2-like protein 1, CACODYLIC ACID, Elongin-B, ... | | Authors: | Olsen, S.K, Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
8FY1
 
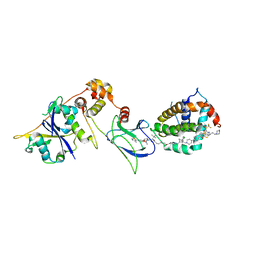 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-2) | | Descriptor: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | Authors: | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
6TWJ
 
 | |
6TWM
 
 | | Product bound structure of the Ectoine utilization protein EutE (DoeB) from Ruegeria pomeroyi | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, ACETATE ION, N-acetyl-L-2,4-diaminobutyric acid deacetylase, ... | | Authors: | Mais, C.-N, Altegoer, F, Bange, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Degradation of the microbial stress protectants and chemical chaperones ectoine and hydroxyectoine by a bacterial hydrolase-deacetylase complex.
J.Biol.Chem., 295, 2020
|
|
6U4Y
 
 | | Crystal Structure of an EZH2-EED Complex in an Oligomeric State | | Descriptor: | Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Jiao, L, Liu, X. | | Deposit date: | 2019-08-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A partially disordered region connects gene repression and activation functions of EZH2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TWK
 
 | | Substrate bound structure of the Ectoine utilization protein EutD (DoeA) from Halomonas elongata | | Descriptor: | (2~{R})-4-azanyl-2-[[(1~{S})-1-oxidanylethyl]amino]butanoic acid, (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Ectoine hydrolase DoeA | | Authors: | Mais, C.-N, Altegoer, F, Bange, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Degradation of the microbial stress protectants and chemical chaperones ectoine and hydroxyectoine by a bacterial hydrolase-deacetylase complex.
J.Biol.Chem., 295, 2020
|
|
