6MJ4
 
 | | Crystal structure of MCD1D/INKTCR TERNARY COMPLEX bound to glycolipid (XXW) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
6UXG
 
 | | Structure of V. metoecus NucC, trimer form | | Descriptor: | SULFATE ION, Vibrio metoecus sp. RC341 NucC | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6MIV
 
 | | Crystal structure of the mCD1d/xxq (JJ300)/iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
6YN1
 
 | | Crystal structure of histone chaperone APLF acidic domain bound to the histone H2A-H2B-H3-H4 octamer | | Descriptor: | Aprataxin and PNK-like factor, CHLORIDE ION, GLYCEROL, ... | | Authors: | Corbeski, I, Guo, X, Van Ingen, H, Sixma, T.K. | | Deposit date: | 2020-04-10 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chaperoning of the histone octamer by the acidic domain of DNA repair factor APLF.
Sci Adv, 8, 2022
|
|
6MJA
 
 | | Crystal structure of the mCD1d/xxo (JJ294) /iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
6YUL
 
 | | CK2 alpha bound to Macrocycle | | Descriptor: | 7,10-Dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.12,6.017,20]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Hanke, T, Kurz, C, Celik, I, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-27 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of pyrazolo[1,5-a]pyrimidines lead to the identification of a highly selective casein kinase 2 inhibitor.
Eur.J.Med.Chem., 208, 2020
|
|
6Z1Z
 
 | | Structure of the anti-CD9 nanobody 4C8 | | Descriptor: | Nanobody 4C8 | | Authors: | Neviani, N, Oosterheert, W, Pearce, N.M, Lutz, M, Kroon-Batenburg, L.M.J, Gros, P. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
6MJQ
 
 | | Crystal structure of the mCD1d/xxp (JJ295) /iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-21 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
6V3W
 
 | | Human Poly(ADP-Ribose) Polymerase 12, Catalytic fragment with four point mutations in complex with RBN-2397 | | Descriptor: | 5-{[(2S)-1-(3-oxo-3-{4-[5-(trifluoromethyl)pyrimidin-2-yl]piperazin-1-yl}propoxy)propan-2-yl]amino}-4-(trifluoromethyl)pyridazin-3(2H)-one, CHLORIDE ION, Protein mono-ADP-ribosyltransferase PARP12 | | Authors: | Swinger, K.K, Gozgit, J.M, Vasbinder, M.M, Wigle, T.J, Kuntz, K.W. | | Deposit date: | 2019-11-26 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | PARP7 negatively regulates the type I interferon response in cancer cells and its inhibition triggers antitumor immunity.
Cancer Cell, 39, 2021
|
|
6V4K
 
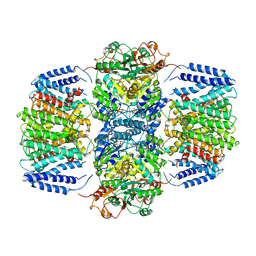 | | Structure of TrkH-TrkA in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Potassium transporter peripheral membrane component, Trk system potassium uptake protein | | Authors: | Zhou, M, Zhang, H. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.53004146 Å) | | Cite: | TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Nat Commun, 11, 2020
|
|
6Z7X
 
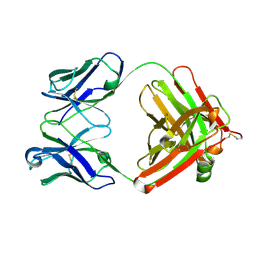 | | Insulin analytical antibody OXI-005 Fab | | Descriptor: | OXI-005 Fab Heavy chain, OXI-005 Fab Light chain | | Authors: | Johansson, E. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insulin binding to the analytical antibody sandwich pair OXI-005 and HUI-018: Epitope mapping and binding properties.
Protein Sci., 30, 2021
|
|
6Z7Y
 
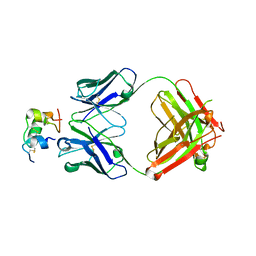 | | Human insulin in complex with the analytical antibody OXI-005 Fab | | Descriptor: | Insulin, OXI-005 Fab Heavy chain, OXI-005 Fab Light chain | | Authors: | Johansson, E. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insulin binding to the analytical antibody sandwich pair OXI-005 and HUI-018: Epitope mapping and binding properties.
Protein Sci., 30, 2021
|
|
6MVH
 
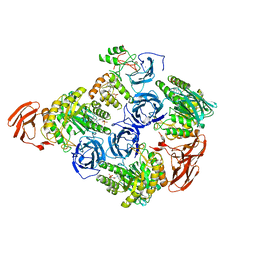 | |
6Z9I
 
 | | Escherichia coli D-2-deoxyribose-5-phosphate aldolase - N21K mutant complex with reaction products | | Descriptor: | 1,2-ETHANEDIOL, Deoxyribose-phosphate aldolase, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Paakkonen, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Substrate specificity of 2-deoxy-D-ribose 5-phosphate aldolase (DERA) assessed by different protein engineering and machine learning methods.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
6Z30
 
 | | Human cation-independent mannose 6-phosphate/ IGF2 receptor domains 9-10 | | Descriptor: | Cation-independent mannose-6-phosphate receptor, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bochel, A.J, Williams, C, Crump, M.P. | | Deposit date: | 2020-05-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7-11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9.
Structure, 28, 2020
|
|
6MJI
 
 | | Crystal structure of the mCD1d/xxs (JJ304) /iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
6B4U
 
 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | Descriptor: | 7-(2-methylphenyl)-1-[2-(morpholin-4-yl)ethyl]-3-{3-[(naphthalen-1-yl)oxy]propyl}-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Judge, R.A, Souers, A.J. | | Deposit date: | 2017-09-27 | | Release date: | 2017-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
6MVL
 
 | |
6Z9J
 
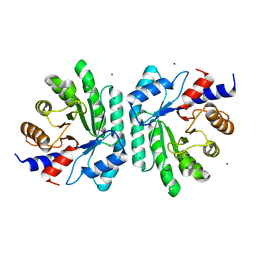 | |
6B5Y
 
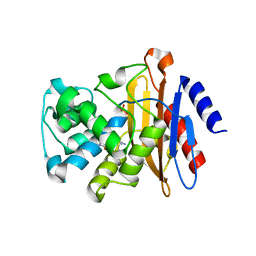 | | Beta-lactamase, mixed with Ceftriaxone, 30ms time point, Shards crystal form | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-09-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6MR8
 
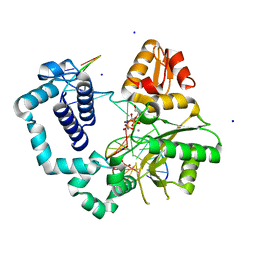 | | D276G DNA polymerase beta substrate complex with templating adenine and incoming Fapy-dGTP analog | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-amino-5-(formylamino)-6-oxo-1,6-dihydropyrimidin-4-yl]-2,5-anhydro-1,3-dideoxy-6-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-D-ribo-hexitol, CALCIUM ION, ... | | Authors: | Freudenthal, B.D, Smith, M.R, Wilson, S.H, Beard, W.A. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | A guardian residue hinders insertion of a Fapy•dGTP analog by modulating the open-closed DNA polymerase transition.
Nucleic Acids Res., 47, 2019
|
|
6B6A
 
 | | Beta-Lactamase, 2secs timepoint, mixed, shards crystal form | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6Z9H
 
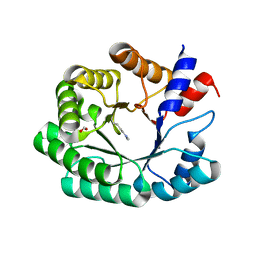 | | Escherichia coli D-2-deoxyribose-5-phosphate aldolase - C47V/G204A/S239D mutant | | Descriptor: | 1,2-ETHANEDIOL, Deoxyribose-phosphate aldolase, FORMIC ACID, ... | | Authors: | Paakkonen, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Substrate specificity of 2-deoxy-D-ribose 5-phosphate aldolase (DERA) assessed by different protein engineering and machine learning methods.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
6B6B
 
 | |
6VH6
 
 | |
