6DCA
 
 | |
6DCB
 
 | | Structure of methylphosphate capping enzyme methyltransferase domain in complex with 5' end of 7SK RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Yang, Y, Eichhorn, C, Cascio, D, Feigon, J. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural basis of 7SK RNA 5'-gamma-phosphate methylation and retention by MePCE.
Nat. Chem. Biol., 15, 2019
|
|
6DCC
 
 | | Structure of methylphosphate capping enzyme methyltransferase domain in complex with 5' end of 7SK RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Yang, Y, Eichhorn, C, Cascio, D, Feigon, J. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of 7SK RNA 5'-gamma-phosphate methylation and retention by MePCE.
Nat. Chem. Biol., 15, 2019
|
|
6DCD
 
 | |
6DCE
 
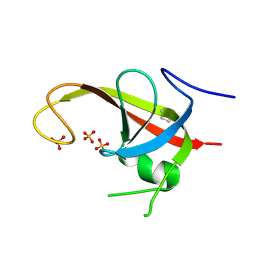 | | X-ray structure of FIP200 claw domain | | Descriptor: | RB1-inducible coiled-coil protein 1, SULFATE ION | | Authors: | Su, M.-Y, Hurley, J.H. | | Deposit date: | 2018-05-05 | | Release date: | 2019-03-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates.
Mol. Cell, 74, 2019
|
|
6DCF
 
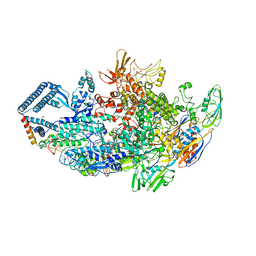 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase and bound to kanglemycin A | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
6DCH
 
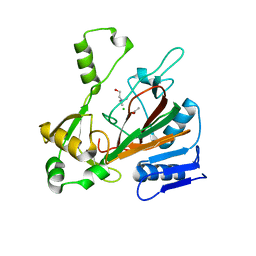 | | Structure of isonitrile biosynthesis enzyme ScoE | | Descriptor: | ACETATE ION, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Born, D.A, Drennan, C.L. | | Deposit date: | 2018-05-07 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isonitrile Formation by a Non-Heme Iron(II)-Dependent Oxidase/Decarboxylase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6DCJ
 
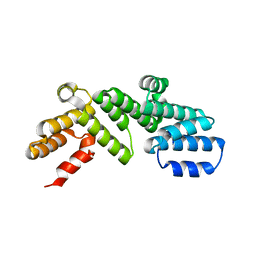 | |
6DCL
 
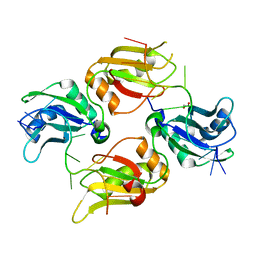 | | Crystal structure of UP1 bound to pri-miRNA-18a terminal loop | | Descriptor: | 1,2-ETHANEDIOL, Heterogeneous nuclear ribonucleoprotein A1, RNA (5'-R(*AP*GP*UP*AP*GP*AP*UP*UP*AP*GP*C)-3') | | Authors: | Kooshapur, H, Sattler, M. | | Deposit date: | 2018-05-07 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structural basis for terminal loop recognition and stimulation of pri-miRNA-18a processing by hnRNP A1.
Nat Commun, 9, 2018
|
|
6DCM
 
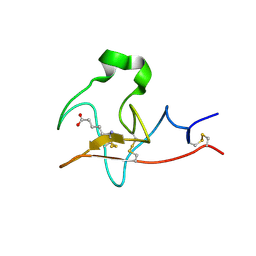 | |
6DCN
 
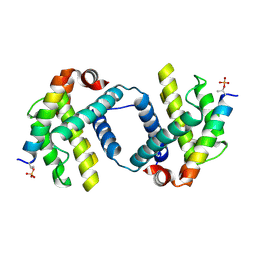 | | Bcl-xL complex with Beclin 1 BH3 domain T108pThr | | Descriptor: | BCL-xl protein, Beclin-1 | | Authors: | Lee, E.F, Smith, B.J, Smith, N.A, Yao, S, Fairlie, W.D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.
Autophagy, 15, 2019
|
|
6DCO
 
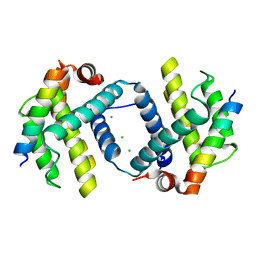 | | Bcl-xL complex with Beclin 1 BH3 domain T108D | | Descriptor: | Bcl-2-like protein 1,Bcl-2-like protein 1, Beclin-1, CHLORIDE ION | | Authors: | Lee, E.F, Smith, B.J, Smith, N.A, Yao, S, Fairlie, W.D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.
Autophagy, 15, 2019
|
|
6DCQ
 
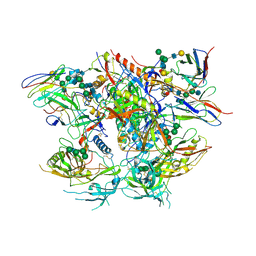 | | Ectodomain of full length, wild type HIV-1 glycoprotein clone PC64M18C043 in complex with PGT151 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rantalainen, K. | | Deposit date: | 2018-05-08 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Co-evolution of HIV Envelope and Apex-Targeting Neutralizing Antibody Lineage Provides Benchmarks for Vaccine Design.
Cell Rep, 23, 2018
|
|
6DCR
 
 | |
6DCS
 
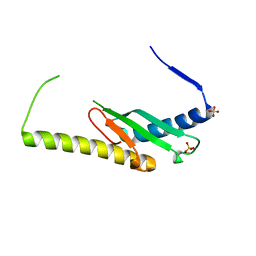 | | Stage III sporulation protein AF (SpoIIIAF) | | Descriptor: | SULFATE ION, Stage III sporulation protein AF | | Authors: | Strynadka, N.C.J, Zeytuni, N, Camp, A.H, Flanagan, K.A. | | Deposit date: | 2018-05-08 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical characterization of SpoIIIAF, a component of a sporulation-essential channel in Bacillus subtilis.
J. Struct. Biol., 204, 2018
|
|
6DCU
 
 | |
6DCV
 
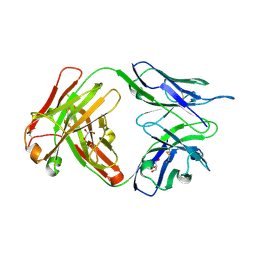 | | Crystal structure of human anti-tau antibody CBTAU-27.1 | | Descriptor: | GLYCEROL, Light chain of CBTAU27.1 Fab, heavy chain of CBTAU-27.1 Fab | | Authors: | Zhu, X, Zhang, H, Wilson, I.A. | | Deposit date: | 2018-05-08 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A common antigenic motif recognized by naturally occurring human VH5-51/VL4-1 anti-tau antibodies with distinct functionalities.
Acta Neuropathol Commun, 6, 2018
|
|
6DCW
 
 | |
6DCX
 
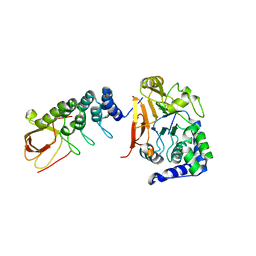 | | iASPP-PP-1c structure and targeting of p53 | | Descriptor: | RelA-associated inhibitor, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Glover, J.N.M, Zhou, Y, Edwards, R.A. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure, 27, 2019
|
|
6DCY
 
 | |
6DCZ
 
 | |
6DD0
 
 | | Crystal structure of VIM-2 complexed with compound 8 | | Descriptor: | ACETATE ION, Beta-lactamase class B VIM-2, ZINC ION, ... | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
6DD1
 
 | |
6DD2
 
 | | Crystal structure of Selaginella moellendorffii HCT | | Descriptor: | Probable hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Lam, C.K, Wang, Y, Weng, J.K. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9056 Å) | | Cite: | Structural and dynamic basis of substrate permissiveness in hydroxycinnamoyltransferase (HCT).
PLoS Comput. Biol., 14, 2018
|
|
