5X4Q
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 5-[[5-chloranyl-2-(pyridin-3-ylmethylamino)pyrimidin-4-yl]amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X9P
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 5 | | Descriptor: | 3-[[4-chloranyl-2-nitro-5-[(2-oxidanylidene-1,3-dihydrobenzimidazol-5-yl)amino]phenyl]amino]propanoic acid, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
5X4P
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 6 | | Descriptor: | 5-[(5-chloranylpyrimidin-4-yl)amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
6CVJ
 
 | |
1Q8J
 
 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima (Cd2+, Hcy, methyltetrahydrofolate complex) | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, ... | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-21 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
6CYF
 
 | | PcrV fragment with bound Fab | | Descriptor: | IgG1 antibody, heavy chain fragment, kappa light chain, ... | | Authors: | Oganesyan, V. | | Deposit date: | 2018-04-05 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Pseudomonas aeruginosa PcrV and Psl, the Molecular Targets of Bispecific Antibody MEDI3902, Are Conserved Among Diverse Global Clinical Isolates.
J. Infect. Dis., 218, 2018
|
|
4XIV
 
 | |
1Q7Z
 
 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima (Cd2+ complex) | | Descriptor: | 5-methyltetrahydrofolate S-homocysteine methyltransferase, CADMIUM ION | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
5H0U
 
 | | Crystal structure of the catalytic domain of membrane type 1 matrix metalloproteinase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Ogata, H, Decaneto, E, Lubitz, W. | | Deposit date: | 2016-10-07 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.239 Å) | | Cite: | Solvent water interactions within the active site of the membrane type I matrix metalloproteinase.
Phys Chem Chem Phys, 19, 2017
|
|
6EC7
 
 | |
5H5Q
 
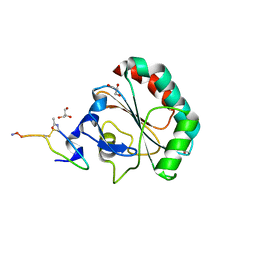 | | Crystal structure of human GPX4 in complex with GXpep-1 | | Descriptor: | GLYCEROL, GXpep-1, Phospholipid hydroperoxide glutathione peroxidase, ... | | Authors: | Sogabe, S, Kadotani, A, Lane, W, Snell, G. | | Deposit date: | 2016-11-09 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery of GPX4 inhibitory peptides from random peptide T7 phage display and subsequent structural analysis
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5H7H
 
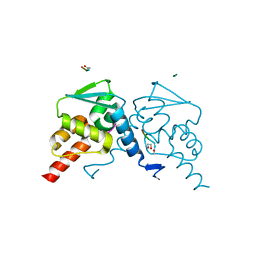 | | Crystal structure of the BCL6 BTB domain in complex with F1324(10-13) | | Descriptor: | 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, F1324 peptide residues 10-13 | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2016-11-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of high-affinity BCL6-binding peptide and its structure-activity relationship.
Biochem. Biophys. Res. Commun., 482, 2017
|
|
4YS0
 
 | |
6EC8
 
 | |
1Q85
 
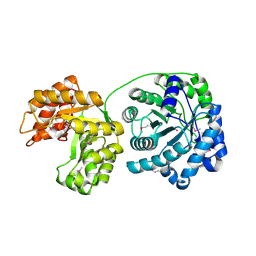 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima (Cd2+ complex, Se-Met) | | Descriptor: | 5-methyltetrahydrofolate S-homocysteine methyltransferase, CADMIUM ION | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-20 | | Release date: | 2004-03-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
5HWZ
 
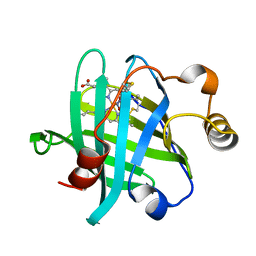 | | Crystal structure of nitrophorin 4 D30N mutant with nitrite | | Descriptor: | NITRITE ION, Nitrophorin-4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ogata, H, He, C, Lubitz, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Elucidation of the heme active site electronic structure affecting the unprecedented nitrite dismutase activity of the ferrihemebproteins, the nitrophorins.
Chem Sci, 7, 2016
|
|
6GMF
 
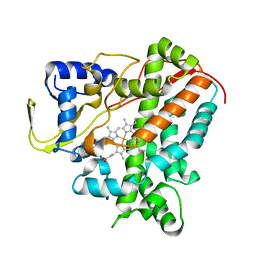 | | Structure of Cytochrome P450 CYP109Q5 from Chondromyces apiculatus | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase | | Authors: | Grogan, G, Dubiel, P, Sharma, M, Klenk, J, Hauer, B. | | Deposit date: | 2018-05-25 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Characterization and structure-guided engineering of the novel versatile terpene monooxygenase CYP109Q5 from Chondromyces apiculatus DSM436.
Microb Biotechnol, 12, 2019
|
|
5H7G
 
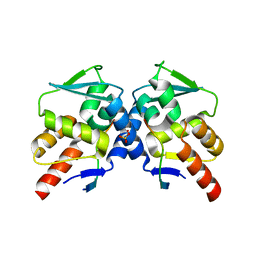 | | Crystal structure of the BCL6 BTB domain in complex with F1324 | | Descriptor: | B-cell lymphoma 6 protein, F1324 peptide, SULFATE ION | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2016-11-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of high-affinity BCL6-binding peptide and its structure-activity relationship.
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5H5R
 
 | | Crystal structure of human GPX4 in complex with GXpep-2 | | Descriptor: | GLYCEROL, GXpep-2, Phospholipid hydroperoxide glutathione peroxidase, ... | | Authors: | Sogabe, S, Kadotani, A, Lane, W, Snell, G. | | Deposit date: | 2016-11-09 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of GPX4 inhibitory peptides from random peptide T7 phage display and subsequent structural analysis
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5H5S
 
 | | Crystal structure of human GPX4 in complex with GXpep-3 | | Descriptor: | GLYCEROL, GXpep-3, Phospholipid hydroperoxide glutathione peroxidase, ... | | Authors: | Sogabe, S, Kadotani, A, Lane, W, Snell, G. | | Deposit date: | 2016-11-09 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of GPX4 inhibitory peptides from random peptide T7 phage display and subsequent structural analysis
Biochem. Biophys. Res. Commun., 482, 2017
|
|
6IAQ
 
 | | Structure of Amine Dehydrogenase from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, Dihydrodipicolinate reductase N-terminus domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Grogan, G, Vaxelaire-Vergne, C, Beloti, L, Mayol, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
1O27
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FAD and BrdUMP at 2.3 A resolution | | Descriptor: | 5-BROMO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
6CVN
 
 | |
1O25
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with dUMP at 2.4 A resolution | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-20 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
1O26
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FAD and dUMP at 1.6 A resolution | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, TRIETHYLENE GLYCOL, ... | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
