3PWA
 
 | | Structure of C126A mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
3D6Q
 
 | | The RNase A- 5'-Deoxy-5'-N-piperidinouridine complex | | Descriptor: | 1-(5-deoxy-5-piperidin-1-yl-alpha-L-arabinofuranosyl)pyrimidine-2,4(1H,3H)-dione, CITRATE ANION, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-05-20 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Morpholino, piperidino, and pyrrolidino derivatives of pyrimidine nucleosides as inhibitors of ribonuclease A: synthesis, biochemical, and crystallographic evaluation.
J.Med.Chem., 52, 2009
|
|
3CNX
 
 | |
6MA7
 
 | | Human CYP3A4 bound to an inhibitor fluconazole | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Cytochrome P450 3A4, ... | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-08-26 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Interaction of Human Drug-Metabolizing CYP3A4 with Small Inhibitory Molecules.
Biochemistry, 58, 2019
|
|
6MAV
 
 | |
3WF6
 
 | | Crystal structure of S6K1 kinase domain in complex with a pyrazolopyrimidine derivative 4-[4-(1H-indol-3-yl)-3,6-dihydropyridin-1(2H)-yl]-1H-pyrazolo[3,4-d]pyrimidine | | Descriptor: | 4-[4-(1H-indol-3-yl)-3,6-dihydropyridin-1(2H)-yl]-1H-pyrazolo[3,4-d]pyrimidine, Ribosomal protein S6 kinase beta-1, ZINC ION | | Authors: | Niwa, H, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Crystal structures of the S6K1 kinase domain in complexes with inhibitors
J.Struct.Funct.Genom., 15, 2014
|
|
3QAG
 
 | | Human Glutathione Transferase O2 with glutathione -new crystal form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Zhou, H, Board, P.G, Oakley, A.J. | | Deposit date: | 2011-01-11 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the dehydroascorbate reductase activity of human omega-class glutathione transferases.
J.Mol.Biol., 420, 2012
|
|
3QEQ
 
 | |
1QHJ
 
 | | X-RAY STRUCTURE OF BACTERIORHODOPSIN GROWN IN LIPIDIC CUBIC PHASES | | Descriptor: | 1,2-[DI-2,6,10,14-TETRAMETHYL-HEXADECAN-16-OXY]-PROPANE, PROTEIN (BACTERIORHODOPSIN), RETINAL | | Authors: | Belrhali, H, Nollert, P, Royant, A, Menzel, C, Rosenbusch, J.P, Landau, E.M, Pebay-Peyroula, E. | | Deposit date: | 1999-05-04 | | Release date: | 1999-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein, lipid and water organization in bacteriorhodopsin crystals: a molecular view of the purple membrane at 1.9 A resolution.
Structure Fold.Des., 7, 1999
|
|
3J3Y
 
 | |
2XF5
 
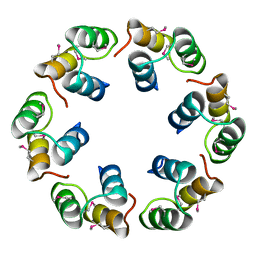 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
1H12
 
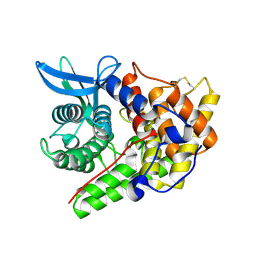 | | Structure of a cold-adapted family 8 xylanase | | Descriptor: | ENDO-1,4-BETA-XYLANASE, alpha-D-xylopyranose, beta-D-xylopyranose | | Authors: | Van Petegem, F, Collins, T, Meuwis, M.A, Feller, G, Gerday, C, Van Beeumen, J. | | Deposit date: | 2002-07-02 | | Release date: | 2003-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Structure of a Cold-Adapted Family 8 Xylanase at 1.3 A Resolution: Structural Adaptations to Cold and Investigation of the Active Site
J.Biol.Chem., 278, 2003
|
|
4BTU
 
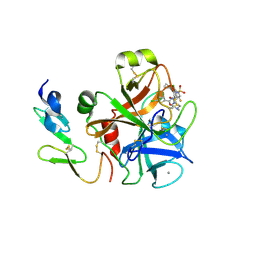 | | Factor Xa in complex with the dual thrombin-FXa inhibitor 57. | | Descriptor: | 5-Chloro-thiophene-2-carboxylic acid [(S)-2-[2-chloro-5-fluoro-3-(2-oxo-piperidin-1-yl)-benzenesulfonylamino]-3-(4-methyl-piperazin-1-yl)-3-oxo-propyl]-amide, CALCIUM ION, COAGULATION FACTOR X HEAVY CHAIN, ... | | Authors: | Meneyrol, J, Follmann, M, Lassalle, G, Wehner, V, Barre, G, Rousseaux, T, Altenburger, J.M, Petit, F, Bocskei, Z, Stehlin-Gaon, C, Schreuder, H, Alet, N, Herault, J.-P, Millet, L, Dol, F, Hasbrand, C, Schaeffer, P, Sadoun, F, Klieber, S, Briot, C, Bono, F, Herbert, J.-M. | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | 5-Chlorothiophene-2-Carboxylic Acid [(S)-2-[2-Methyl-3-(2-Oxopyrrolidin-1-Yl)Benzenesulfonylamino]-3-(4-Methylpiperazin-1-Yl)-3-Oxopropyl]Amide (Sar107375), a Selective and Potent Orally Active Dual Thrombin and Factor Xa Inhibitor.
J.Med.Chem., 56, 2013
|
|
2XSC
 
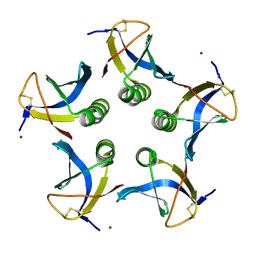 | | Crystal structure of the cell-binding B oligomer of verotoxin-1 from E. coli | | Descriptor: | SHIGA-LIKE TOXIN 1 SUBUNIT B, ZINC ION | | Authors: | Stein, P.E, Boodhoo, A, Tyrrell, G.J, Brunton, J.L, Oeffner, R.D, Bunkoczi, G, Read, R.J. | | Deposit date: | 2010-09-27 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal Structure of the Cell-Binding B Oligomer of Verotoxin-1 from E. Coli.
Nature, 355, 1992
|
|
7B12
 
 | | HUMAN IMMUNOPROTEASOME 20S PARTICLE IN COMPLEX WITH [2-(3-ethylphenyl)-1-[(2S)-3-phenyl-2-[(pyrazin-2-yl)formamido]propanamido]ethyl]boronic acid | | Descriptor: | ((R)-2-(3-ethylphenyl)-1-((S)-3-phenyl-2-(pyrazine-2-carboxamido)propanamido)ethyl)boronic acid, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Musil, D, Klein, M, Crosignani, S. | | Deposit date: | 2020-11-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure-Based Optimization and Discovery of M3258, a Specific Inhibitor of the Immunoproteasome Subunit LMP7 ( beta 5i).
J.Med.Chem., 64, 2021
|
|
1HRH
 
 | |
4P36
 
 | | Crystal structure of DJ-1 With Zn(II) bound (crystal 2) | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, Protein DJ-1, ZINC ION | | Authors: | Tashiro, S, Wu, C.-X, Hoang, Q.Q, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.182 Å) | | Cite: | Thermodynamic and Structural Characterization of the Specific Binding of Zn(II) to Human Protein DJ-1.
Biochemistry, 53, 2014
|
|
2OR3
 
 | |
3W59
 
 | |
2W6K
 
 | | The crystal structure at 1.7 A resolution of CobE, a protein from the cobalamin (vitamin B12) biosynthetic pathway | | Descriptor: | COBE, GLYCEROL, SULFATE ION | | Authors: | Vevodova, J, Smith, D, McGoldrick, H, Deery, E, Murzin, A.G, Warren, M.J, Wilson, K.S. | | Deposit date: | 2008-12-18 | | Release date: | 2008-12-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure at 1.7 A Resolution of Cobe, a Protein from the Cobalamin (Vitamin B12) Biosynthetic Pathway
To be Published
|
|
4BWE
 
 | | Crystal structure of C-terminally truncated glypican-1 after controlled dehydration to 86 percent relative humidity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Glypican-1 | | Authors: | Awad, W, Svensson Birkedal, G, Thunnissen, M.M.G.M, Mani, K, Logan, D.T. | | Deposit date: | 2013-07-01 | | Release date: | 2013-12-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Improvements in the order, isotropy and electron density of glypican-1 crystals by controlled dehydration.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
4KRF
 
 | |
7EW1
 
 | | Cryo-EM structure of siponimod -bound Sphingosine-1-phosphate receptor 5 in complex with Gi protein | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Y, Jia, G.W, Shao, Z.H, Su, Z.M. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
7EW4
 
 | | Cryo-EM structure of CYM-5541-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
7EVY
 
 | | Cryo-EM structure of siponimod -bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jia, G.W, Yuan, Y, Su, Z.M, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
