7SHB
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI79 | | Descriptor: | 3C-like proteinase nsp5, benzyl [(2S)-1-({(2S)-1-[2-(3-amino-3-oxopropyl)-2-propanoylhydrazinyl]-4-methyl-1-oxopentan-2-yl}amino)-3-methyl-1-oxobutan-2-yl]carbamate (non-preferred name) | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2021-10-08 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the SARS-CoV-2 main protease in complex with inhibitors
To Be Published
|
|
6U4Q
 
 | | Carbonic anhydrase 2 in complex with SB4197 | | Descriptor: | 4-methyl-1lambda~6~,2,4-benzothiadiazine-1,1,3(2H,4H)-trione, Carbonic anhydrase 2, ZINC ION | | Authors: | Murray, A.B, Lomelino, C.L, McKenna, R. | | Deposit date: | 2019-08-26 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.306 Å) | | Cite: | "A Sweet Combination": Developing Saccharin and Acesulfame K Structures for Selectively Targeting the Tumor-Associated Carbonic Anhydrases IX and XII.
J.Med.Chem., 63, 2020
|
|
6OE0
 
 | |
7NI1
 
 | | CRYSTAL STRUCTURE OF NATIVE HUMAN MYELOPEROXIDASE IN COMPLEX WITH CPD 9 | | Descriptor: | (S)-1-(2-(amino(phenyl)methyl)benzyl)-2-thioxo-1,2,3,5-tetrahydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sjogren, T, Inghardt, T. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of AZD4831, a Mechanism-Based Irreversible Inhibitor of Myeloperoxidase, As a Potential Treatment for Heart Failure with Preserved Ejection Fraction.
J.Med.Chem., 65, 2022
|
|
6OE4
 
 | |
8C85
 
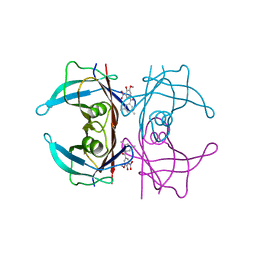 | | Crystal structure of human transthyretin in complex with 3-O-methyltolcapone analogue 1 | | Descriptor: | (3,5-dimethylphenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | Authors: | Poonsiri, T, Benini, S, Loconte, V, Cianci, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | 3-O-Methyltolcapone and Its Lipophilic Analogues Are Potent Inhibitors of Transthyretin Amyloidogenesis with High Permeability and Low Toxicity.
Int J Mol Sci, 25, 2023
|
|
6M7F
 
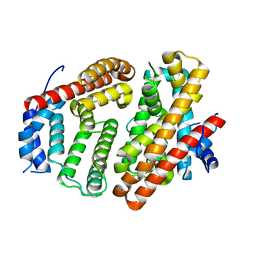 | | Wild-type Cucumene Synthase | | Descriptor: | Cucumene Synthase | | Authors: | Blank, P.N, Pemberton, T.A, Christianson, D.W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.051 Å) | | Cite: | Crystal Structure of Cucumene Synthase, a Terpenoid Cyclase That Generates a Linear Triquinane Sesquiterpene.
Biochemistry, 57, 2018
|
|
7NGI
 
 | |
7NGY
 
 | |
7NFL
 
 | |
7V52
 
 | | Structure of AdaV | | Descriptor: | AdaV, FE (III) ION | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
6U0J
 
 | | Crosslinked Crystal Structure of Malonyl-CoA Acyl Carrier Protein Transacylase, FabD, and Acyl Carrier Protein, AcpP | | Descriptor: | 1,2-ETHANEDIOL, Acyl carrier protein, Malonyl CoA-acyl carrier protein transacylase, ... | | Authors: | Mindrebo, J.T, Misson, L, Davis, T.D, Burkart, M.D, Noel, J.P. | | Deposit date: | 2019-08-14 | | Release date: | 2020-07-22 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interfacial plasticity facilitates high reaction rate of E. coli FAS malonyl-CoA:ACP transacylase, FabD.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7NGW
 
 | |
6OEZ
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor (+)-N-(Cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrro-lidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, N-(cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrrolidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
7NLC
 
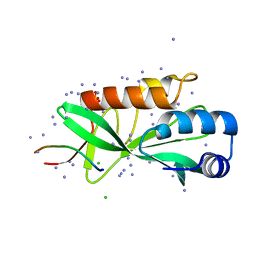 | | Crystallographic structure of human Tsg101 UEV domain in complex with a HEV ORF3 peptide | | Descriptor: | AMMONIUM ION, CHLORIDE ION, Protein ORF3, ... | | Authors: | Moschidi, D, Dupre, E, Villeret, V, Hanoulle, X. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Crystallographic structure of human Tsg101 UEV domain in complex with a HEV ORF3 peptide
To Be Published
|
|
7V7M
 
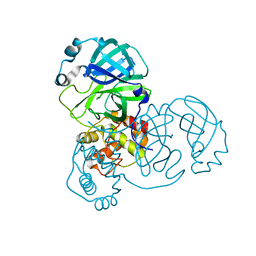 | | crystal structure of SARS-CoV-2 3CL protease | | Descriptor: | 3C-like proteinase | | Authors: | Yi, Y, Zhang, M, Ye, M. | | Deposit date: | 2021-08-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Schaftoside inhibits 3CL pro and PL pro of SARS-CoV-2 virus and regulates immune response and inflammation of host cells for the treatment of COVID-19.
Acta Pharm Sin B, 12, 2022
|
|
7V57
 
 | | Structure of AdaV | | Descriptor: | 2-OXOGLUTARIC ACID, AdaV, CHLORIDE ION, ... | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
7NFA
 
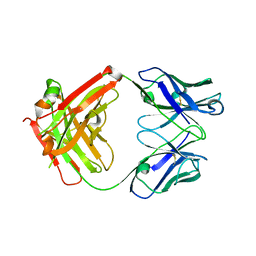 | | The structure of the humanised A33 Fab C226S variant, an immunotherapy candidate for colorectal cancer | | Descriptor: | A33 Fab heavy chain, A33 Fab light chain | | Authors: | Tang, J, Zhang, C, Dalby, P, Kozielski, F. | | Deposit date: | 2021-02-05 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the humanised A33 Fab C226S variant, an immunotherapy candidate for colorectal cancer
To Be Published
|
|
8C9L
 
 | |
6OKN
 
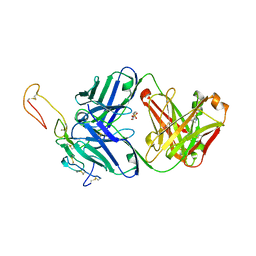 | | OX40R (TNFRSF4) bound to Fab 1A7 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab 1A7 heavy chain, Fab 1A7 light chain, ... | | Authors: | Ultsch, M.H, Boenig, G, Harris, S.F. | | Deposit date: | 2019-04-14 | | Release date: | 2019-07-10 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Tetravalent biepitopic targeting enables intrinsic antibody agonism of tumor necrosis factor receptor superfamily members.
Mabs, 11, 2019
|
|
5AJS
 
 | |
8C62
 
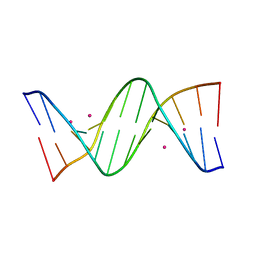 | | Crystal structure of cisplatin/B-DNA adduct | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PLATINUM (II) ION | | Authors: | Troisi, R, Tito, G, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-01-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
8C9O
 
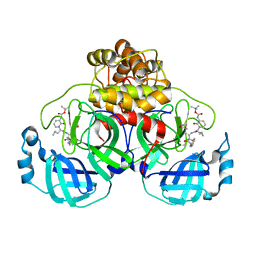 | | Crystal structure of SARS-CoV-2 Mpro-S144A mutant in complex with 13b-K | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2023-01-23 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of SARS-CoV-2 Mpro_S144A mutant in complex with 13b-K
To Be Published
|
|
5AF7
 
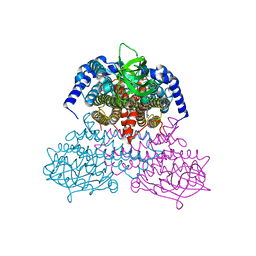 | | 3-Sulfinopropionyl-coenzyme A (3SP-CoA) desulfinase from Advenella mimigardefordensis DPN7T: crystal structure and function of a desulfinase with an acyl-CoA dehydrogenase fold. Native crystal structure | | Descriptor: | ACYL-COA DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cianci, M, Schuermann, M, Meijers, R, Schneider, T.R, Steinbuechel, A. | | Deposit date: | 2015-01-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | 3-Sulfinopropionyl-Coenzyme a (3Sp-Coa) Desulfinase from Advenella Mimigardefordensis Dpn7(T): Crystal Structure and Function of a Desulfinase with an Acyl-Coa Dehydrogenase Fold.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5AD3
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | 3-methoxy-N-[2-[4-[1-(3-methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phenoxy]ethyl]-N-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
