6MFR
 
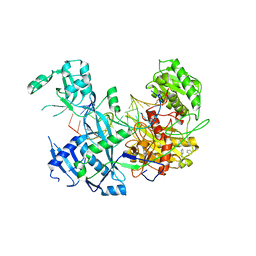 | |
6MFN
 
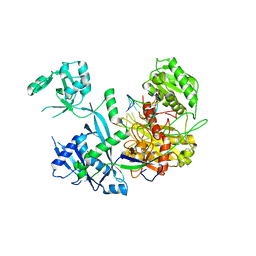 | |
6NIT
 
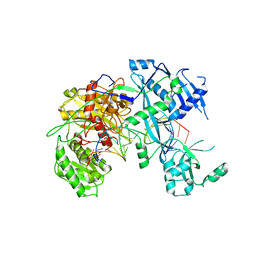 | |
5M9D
 
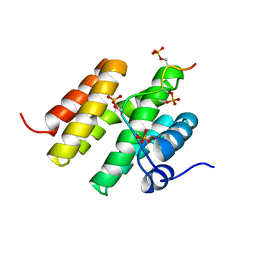 | |
7OM7
 
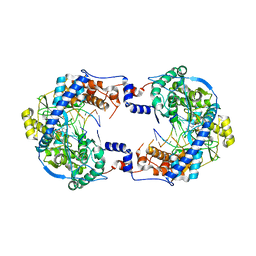 | | Thosea asigna virus RdRP domain in complex with RNA and nucleotide UMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Ferrero, D.S, Falqui, M, Verdaguer, N. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Snapshots of a Non-Canonical RdRP in Action.
Viruses, 13, 2021
|
|
3OTC
 
 | |
4Z4D
 
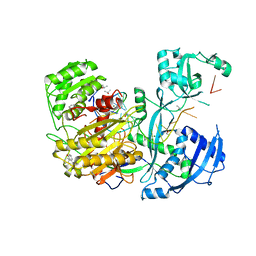 | | Human Argonaute2 Bound to t1-G Target RNA | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, PHENOL, ... | | Authors: | Schirle, N.T, MacRae, I.J. | | Deposit date: | 2015-04-02 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets.
Elife, 4, 2015
|
|
4Z4F
 
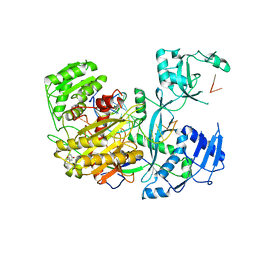 | | Human Argonaute2 Bound to t1-DAP Target RNA | | Descriptor: | MAGNESIUM ION, PHENOL, Protein argonaute-2, ... | | Authors: | Schirle, N.T, MacRae, I.J. | | Deposit date: | 2015-04-02 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets.
Elife, 4, 2015
|
|
4Z4H
 
 | |
4Z4I
 
 | |
4Z4C
 
 | | Human Argonaute2 Bound to t1-C Target RNA | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, PHENOL, ... | | Authors: | Schirle, N.T, MacRae, I.J. | | Deposit date: | 2015-04-02 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets.
Elife, 4, 2015
|
|
4Z4G
 
 | |
4Z4E
 
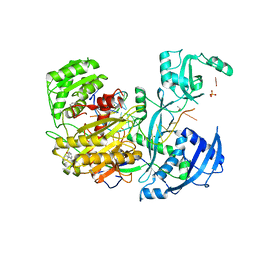 | | Human Argonaute2 Bound to t1-U Target RNA | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, PHENOL, ... | | Authors: | Schirle, N.T, MacRae, I.J. | | Deposit date: | 2015-04-02 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets.
Elife, 4, 2015
|
|
6K32
 
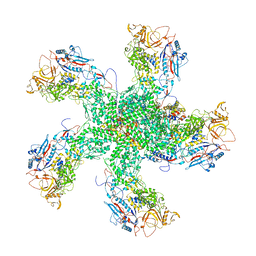 | | RdRp complex | | Descriptor: | 2'-O-methyladenosine 5'-(dihydrogen phosphate), 7-METHYLGUANOSINE, DIPHOSPHATE, ... | | Authors: | Li, X.W. | | Deposit date: | 2019-05-16 | | Release date: | 2019-11-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of RdRps Within a Transcribing dsRNA Virus Provides Insights Into the Mechanisms of RNA Synthesis.
J.Mol.Biol., 432, 2020
|
|
5OMQ
 
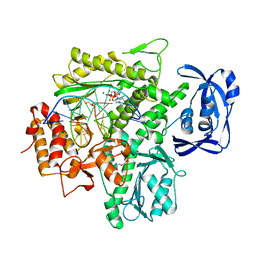 | | Ternary complex of 9N DNA polymerase in the replicative state with three metal ions in the active site | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA Primer, DNA Template, ... | | Authors: | Betz, K, Marx, A, Diederichs, K. | | Deposit date: | 2017-08-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Crystal structures of ternary complexes of archaeal B-family DNA polymerases.
PLoS ONE, 12, 2017
|
|
5OMV
 
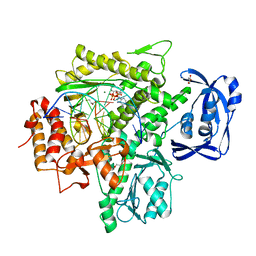 | | Ternary complex of 9N DNA polymerase in the replicative state with two metal ions in the active site | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA polymerase, DNA primer, ... | | Authors: | Betz, K, Marx, A, Diederichs, K. | | Deposit date: | 2017-08-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Crystal structures of ternary complexes of archaeal B-family DNA polymerases.
PLoS ONE, 12, 2017
|
|
4OI4
 
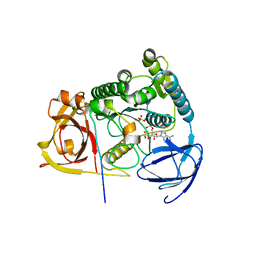 | | Protein complex of Clp1 bound to ATP and Mg2+ with Pcf11deltaN454deltaC563 of S. cerevisiae | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Dikfidan, A, Loll, B, Zeymer, C, Clausen, T, Meinhart, A. | | Deposit date: | 2014-01-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | RNA specificity and regulation of catalysis in the eukaryotic polynucleotide kinase clp1.
Mol.Cell, 54, 2014
|
|
8B9I
 
 | |
8B9G
 
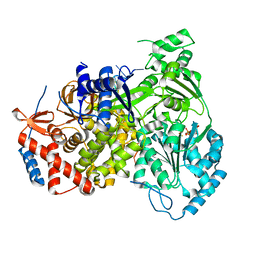 | | Cryo-EM structure of MLE in complex with ADP:AlF4 and U10 RNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dosage compensation regulator, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis of RNA-induced autoregulation of the DExH-type RNA helicase maleless.
Mol.Cell, 83, 2023
|
|
3JSB
 
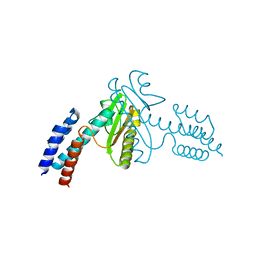 | | Crystal structure of the N-terminal domain of the Lymphocytic Choriomeningitis Virus L protein | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Morin, B, Jamal, S, Ferron, F.P, Coutard, B, Bricogne, G, Canard, B, Vonrhein, C, Marseilles Structural Genomics Program @ AFMB (MSGP) | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription
Plos Pathog., 6, 2010
|
|
7QDY
 
 | | RNA-bound human SKI complex | | Descriptor: | Helicase SKI2W, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'), Tetratricopeptide repeat protein 37, ... | | Authors: | Koegel, A, Keidel, A, Bonneau, F, Schaefer, I.B, Conti, E. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism.
Mol.Cell, 82, 2022
|
|
8PJB
 
 | |
8PJJ
 
 | |
3LLM
 
 | | Crystal Structure Analysis of a RNA Helicase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase A, CACODYLATE ION, ... | | Authors: | Schutz, P, Karlberg, T, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human RNA helicase A (DHX9): structural basis for unselective nucleotide base binding in a DEAD-box variant protein.
J.Mol.Biol., 400, 2010
|
|
4L87
 
 | |
