4E4D
 
 | | Crystal structure of mouse RANKL-OPG complex | | Descriptor: | CHLORIDE ION, Tumor necrosis factor ligand superfamily member 11, soluble form, ... | | Authors: | Nelson, C.A, Fremont, D.H. | | Deposit date: | 2012-03-12 | | Release date: | 2012-10-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RANKL Employs Distinct Binding Modes to Engage RANK and the Osteoprotegerin Decoy Receptor.
Structure, 20, 2012
|
|
3QD6
 
 | | Crystal structure of the CD40 and CD154 (CD40L) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD40 ligand, Tumor necrosis factor receptor superfamily member 5 | | Authors: | Lee, J.-O, Kim, Y.J, Song, D.H, Kim, H.M, Park, B.S. | | Deposit date: | 2011-01-18 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation
J.Biol.Chem., 286, 2011
|
|
5BNQ
 
 | | Crystal structure of hRANKL-mRANK complex | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Ren, J. | | Deposit date: | 2015-05-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A RANKL mutant used as an inter-species vaccine for efficient immunotherapy of osteoporosis.
Sci Rep, 5, 2015
|
|
3K51
 
 | | Crystal Structure of DcR3-TL1A complex | | Descriptor: | Decoy receptor 3, Tumor necrosis factor ligand superfamily member 15, secreted form | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2009-10-06 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
5CIR
 
 | | Crystal structure of death receptor 4 (DR4; TNFFRSF10A) bound to TRAIL (TNFSF10) | | Descriptor: | CHLORIDE ION, Tumor necrosis factor ligand superfamily member 10, Tumor necrosis factor receptor superfamily member 10A, ... | | Authors: | Sheriff, S. | | Deposit date: | 2015-07-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the death receptor 4-TNF-related apoptosis-inducing ligand (DR4-TRAIL) complex.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4GIQ
 
 | | Crystal Structure of mouse RANK bound to RANKL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SODIUM ION, ... | | Authors: | Nelson, C.A, Wang, M.W.-H, Fremont, D.H. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RANKL Employs Distinct Binding Modes to Engage RANK and the Osteoprotegerin Decoy Receptor.
Structure, 20, 2012
|
|
6MGP
 
 | | Structure of human 4-1BB / 4-1BBL complex | | Descriptor: | ACETATE ION, GLYCEROL, Tumor necrosis factor ligand superfamily member 9, ... | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab
Nat Commun, 9, 2018
|
|
6MKZ
 
 | | Crystal structure of murine 4-1BB/4-1BBL complex | | Descriptor: | Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bitra, A, Zajonc, D.M, Doukov, T. | | Deposit date: | 2018-09-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the m4-1BB/4-1BBL complex reveals an unusual dimeric ligand that undergoes structural changes upon 4-1BB receptor binding.
J. Biol. Chem., 294, 2019
|
|
4J6G
 
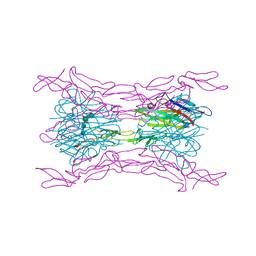 | | CRYSTAL STRUCTURE OF LIGHT AND DcR3 COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Liu, W, Zhan, C, Bonanno, J.B, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-11 | | Release date: | 2013-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
3ALQ
 
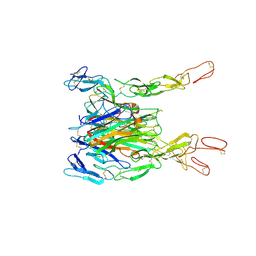 | | Crystal structure of TNF-TNFR2 complex | | Descriptor: | COBALT (II) ION, Tumor necrosis factor, Tumor necrosis factor receptor superfamily member 1B | | Authors: | Mukai, Y, Nakamura, T, Yamagata, Y, Tsutsumi, Y. | | Deposit date: | 2010-08-06 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Solution of the Structure of the TNF-TNFR2 Complex
Sci.Signal., 3, 2010
|
|
5L36
 
 | |
1D4V
 
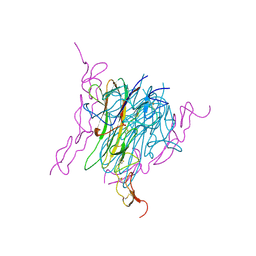 | | Crystal structure of trail-DR5 complex | | Descriptor: | DEATH RECEPTOR 5, TNF-RELATED APOPTOSIS INDUCING LIGAND | | Authors: | Mongkolsapaya, J, Grimes, J.M, Stuart, D.I, Jones, E.Y, Screaton, G.R. | | Deposit date: | 1999-10-06 | | Release date: | 1999-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the TRAIL-DR5 complex reveals mechanisms conferring specificity in apoptotic initiation
Nat.Struct.Biol., 6, 1999
|
|
6A3V
 
 | | Complex structure of human 4-1BB and 4-1BBL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9 | | Authors: | Li, Y, Zhang, C, Chai, Y, Qi, J, Tien, P, Gao, S, Gao, G.F. | | Deposit date: | 2018-06-17 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.391 Å) | | Cite: | Limited Cross-Linking of 4-1BB by 4-1BB Ligand and the Agonist Monoclonal Antibody Utomilumab.
Cell Rep, 25, 2018
|
|
1D0G
 
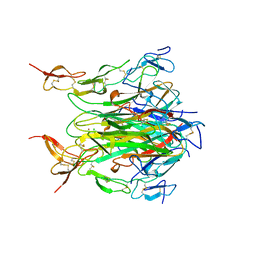 | | CRYSTAL STRUCTURE OF DEATH RECEPTOR 5 (DR5) BOUND TO APO2L/TRAIL | | Descriptor: | APOPTOSIS-2 LIGAND, CHLORIDE ION, DEATH RECEPTOR-5, ... | | Authors: | Hymowitz, S.G, Christinger, H.W, Fuh, G, O'Connell, M.P, Kelley, R.F, Ashkenazi, A, de Vos, A.M. | | Deposit date: | 1999-09-09 | | Release date: | 1999-10-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Triggering cell death: the crystal structure of Apo2L/TRAIL in a complex with death receptor 5.
Mol.Cell, 4, 1999
|
|
1TNR
 
 | |
1DU3
 
 | | Crystal structure of TRAIL-SDR5 | | Descriptor: | DEATH RECEPTOR 5, TNF-RELATED APOPTOSIS INDUCING LIGAND, ZINC ION | | Authors: | Cha, S.-S, Sung, B.-J, Oh, B.-H. | | Deposit date: | 2000-01-14 | | Release date: | 2000-09-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TRAIL-DR5 complex identifies a critical role of the unique frame insertion in conferring recognition specificity
J.Biol.Chem., 275, 2000
|
|
6BRB
 
 | | Novel non-antibody protein scaffold targeting CD40L | | Descriptor: | CD40 ligand, Tn3-like, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Oganesyan, V, Baca, M, Thisted, T, Grinberg, L, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2017-11-30 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | A CD40L-targeting protein reduces autoantibodies and improves disease activity in patients with autoimmunity.
Sci Transl Med, 11, 2019
|
|
6BWV
 
 | | Crystal Structure of the 4-1BB/4-1BBL Complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9 | | Authors: | Oganesyan, V, Gilbreth, R.N, Baca, M. | | Deposit date: | 2017-12-15 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human 4-1BB/4-1BBL complex.
J. Biol. Chem., 293, 2018
|
|
6CPR
 
 | | Crystal structure of 4-1BBL/4-1BB complex in C2 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Aruna, B, Zajonc, D.M. | | Deposit date: | 2018-03-14 | | Release date: | 2018-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the human 4-1BB receptor bound to its ligand 4-1BBL reveal covalent receptor dimerization as a potential signaling amplifier.
J. Biol. Chem., 293, 2018
|
|
4MSV
 
 | | Crystal structure of FASL and DcR3 complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 6, ... | | Authors: | Liu, W, Ramagopal, U.A, Zhan, C, Bonanno, J.B, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-18 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Complex of Human FasL and Its Decoy Receptor DcR3.
Structure, 24, 2016
|
|
4N90
 
 | |
4KGQ
 
 | | Crystal structure of a human light loop mutant in complex with dcr3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 14, ... | | Authors: | Liu, W, Zhan, C, Bonanno, J.B, Sampathkumar, P, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-04-29 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
4MXW
 
 | | Structure of heterotrimeric lymphotoxin LTa1b2 bound to lymphotoxin beta receptor LTbR and anti-LTa Fab | | Descriptor: | Lymphotoxin-alpha, Lymphotoxin-beta, Tumor necrosis factor receptor superfamily member 3, ... | | Authors: | Sudhamsu, J, Yin, J.P, Hymowitz, S.G. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Dimerization of LT beta R by LT alpha 1 beta 2 is necessary and sufficient for signal transduction.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KGG
 
 | | Crystal structure of light mutant2 and dcr3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 14, ... | | Authors: | Liu, W, Bonanno, J.B, Zhan, C, Kumar, P.R, Toro, R, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-29 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
7KP7
 
 | | asymmetric mTNF-alpha hTNFR1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Tumor necrosis factor, ... | | Authors: | Arakaki, T.L, Fox III, D, Edwards, T.E, Foley, A, Ceska, T. | | Deposit date: | 2020-11-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into the disruption of TNF-TNFR1 signalling by small molecules stabilising a distorted TNF.
Nat Commun, 12, 2021
|
|
