6QRS
 
 | | X-ray radiation dose series on xylose isomerase - 0.13 MGy | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Taberman, H, Bury, C.S, van der Woerd, M.J, Snell, E.H, Garman, E.F. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural knowledge or X-ray damage? A case study on xylose isomerase illustrating both.
J.Synchrotron Radiat., 26, 2019
|
|
8DUN
 
 | | Cryo-EM Structure of Antibody SKW11 in complex with Western Equine Encephalitis Virus spike (local refinement from VLP particles) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody SKW11 heavy chain, ... | | Authors: | Cerutti, G, Verardi, R, Roederer, M, Shapiro, L. | | Deposit date: | 2022-07-27 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (5.84 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
7RRF
 
 | | Carbonic Anhydrase IX-mimic in complex with Beta-Galactose_2C | | Descriptor: | 2-[1-(1,1,3-trioxo-2,3-dihydro-1H-1lambda~6~,2-benzothiazol-6-yl)-1H-1,2,3-triazol-4-yl]ethyl beta-L-gulopyranoside, Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, McKenna, R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Inhibition of Carbonic Anhydrase IX and Monocarboxylate Transporters 1 and 4 in breast cancer via novel inhibition with Beta-Galactose 2C
To Be Published
|
|
8DK4
 
 | | Peroxisome proliferator-activated receptor gamma in complex with VSP-51-2 | | Descriptor: | 5-(benzylcarbamoyl)-1-[(4-chloro-3-fluorophenyl)methyl]-1H-indole-2-carboxylic acid, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Ma, L, Zhou, X.E, Suino-Powell, K, Hou, N, Zhou, Z, Luo, J, Xu, H.E, Yi, W. | | Deposit date: | 2022-07-02 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of VSP-51-2 as the Novel and Safe PPAR gamma Modulator: Structure-Based Design, Biological Validation and Crystal Analysis
To Be Published
|
|
8UQO
 
 | | PLCb3-Gbg-Gaq complex on membranes | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | The mechanism of G alpha q regulation of PLC beta 3 -catalyzed PIP2 hydrolysis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6Y4B
 
 | |
7UBB
 
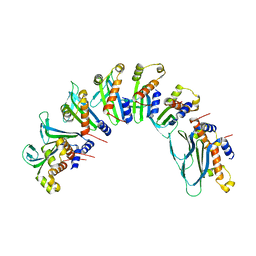 | |
8QDJ
 
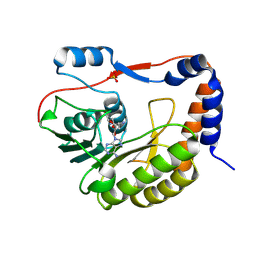 | |
8E71
 
 | | Staphylococcus aureus ClpP in complex with compound 3471 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5S,6S,9aS)-N-[(4-fluorophenyl)methyl]-6-methyl-8-[(naphthalen-1-yl)methyl]-4,7-dioxohexahydro-2H-pyrazino[1,2-a]pyrimidine-1(6H)-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2022-08-23 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Development of a high throughput and site specific, fluorescent polarization assay to screen for activators of Caseinolytic Protease P leads to the discovery of synthetically tractable new activator class
To Be Published
|
|
8QPZ
 
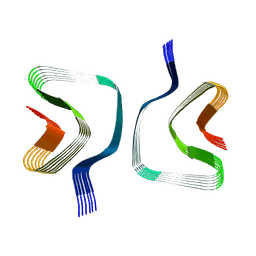 | | CryoEM structure of recombinant DeltaN7 alpha-synuclein in PBS | | Descriptor: | Alpha-synuclein | | Authors: | Thacker, D, Wilkinson, M, Dewison, K.M, Ranson, N.A, Brockwell, D.J, Radford, S.E. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Residues 2 to 7 of alpha-synuclein regulate amyloid formation via lipid-dependent and lipid-independent pathways.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6SSP
 
 | | Structure of the pentameric ligand-gated ion channel ELIC in complex with a NAM nanobody | | Descriptor: | CALCIUM ION, Cys-loop ligand-gated ion channel, NANOBODY 21, ... | | Authors: | Ulens, C, Brams, M, Evans, G.L, Spurny, R, Govaerts, C, Pardon, E, Steyaert, J. | | Deposit date: | 2019-09-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Modulation of the Erwinia ligand-gated ion channel (ELIC) and the 5-HT 3 receptor via a common vestibule site.
Elife, 9, 2020
|
|
8DNG
 
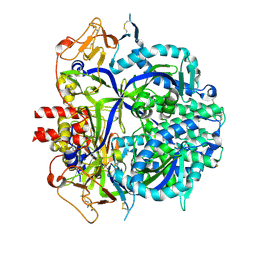 | |
6SP4
 
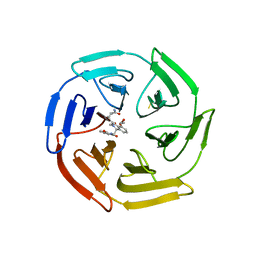 | | KEAP1 IN COMPLEX WITH COMPOUND 23 | | Descriptor: | (1~{S},2~{R})-2-[[(1~{S})-1-[[1,3-bis(oxidanylidene)isoindol-2-yl]methyl]-5-(2-hydroxyethyloxy)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]cyclobutane-1-carboxamide, Kelch-like ECH-associated protein 1 | | Authors: | Ontoria, J.M, Biancofiore, I, Fezzardi, P, Torrente de Haro, E, Colarusso, S, Bianchi, E, Andreini, M, Patsilinakos, A, Summa, V, Pacifici, R, Munoz-Sanjuan, I, Park, L, Bresciani, A, Dominguez, C, Toledo-Sherman, L, Harper, S. | | Deposit date: | 2019-08-30 | | Release date: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Combined Peptide and Small-Molecule Approach toward Nonacidic THIQ Inhibitors of the KEAP1/NRF2 Interaction.
Acs Med.Chem.Lett., 11, 2020
|
|
8DNR
 
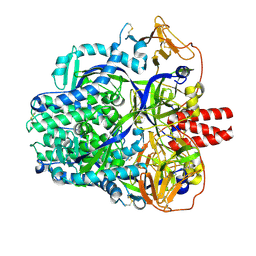 | |
5MQT
 
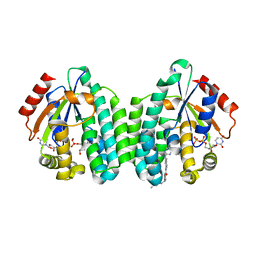 | | Crystal structure of dCK mutant C3S in complex with imatinib and UDP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Deoxycytidine kinase, ... | | Authors: | Saez-Ayala, M, Rebuffet, E, Hammam, K, Gros, L, Lopez, S, Hajem, B, Humbert, M, Baudelet, E, Audebert, S, Betzi, S, Lugari, A, Combes, S, Pez, D, Letard, S, Mansfield, C, Moussy, A, de Sepulveda, P, Morelli, X, Dubreuil, P. | | Deposit date: | 2016-12-20 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dual protein kinase and nucleoside kinase modulators for rationally designed polypharmacology.
Nat Commun, 8, 2017
|
|
8E9A
 
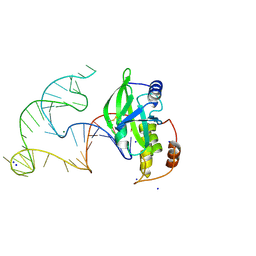 | | Crystal structure of AsfvPolX in complex with 10-23 DNAzyme and Mg | | Descriptor: | DNA/RNA (52-MER), MAGNESIUM ION, Repair DNA polymerase X, ... | | Authors: | Cramer, E.R, Robart, A.R, Kaya, A.I. | | Deposit date: | 2022-08-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of a 10-23 deoxyribozyme exhibiting a homodimer conformation.
Commun Chem, 6, 2023
|
|
6XH1
 
 | |
8PAZ
 
 | | OXIDIZED NATIVE PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-24 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
8DO4
 
 | |
5MX5
 
 | | Mouse PA28alpha-beta | | Descriptor: | PHOSPHATE ION, Proteasome activator complex subunit 1, Proteasome activator complex subunit 2 | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2017-01-20 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Mammalian Proteasome Activator PA28 Forms an Asymmetric alpha 4 beta 3 Complex.
Structure, 25, 2017
|
|
8DWO
 
 | | Cryo-EM Structure of Eastern Equine Encephalitis Virus in complex with SKE26 Fab | | Descriptor: | Envelope glycoprotein E1, Envelope glycoprotein E2, SKE26 Fab Heavy Chain, ... | | Authors: | Pletnev, S, Verardi, R, Roedeger, M, Kwong, P. | | Deposit date: | 2022-08-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
5M95
 
 | | STAPHYLOCOCCUS CAPITIS DIVALENT METAL ION TRANSPORTER (DMT) IN COMPLEX WITH MANGANESE | | Descriptor: | CAMELID ANTIBODY FRAGMENT, NANOBODY, Divalent metal cation transporter MntH, ... | | Authors: | Ehrnstorfer, I.A, Geertsma, E.R, Pardon, E, Steyaert, J, Dutzler, R. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure Of A Slc11 (Nramp) Transporter Reveals The Basis For Transition-Metal Ion Transport.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7R36
 
 | | Difference-refined structure of fatty acid photodecarboxylase 2 microsecond following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R33
 
 | | Difference-refined structure of fatty acid photodecarboxylase 20 ps following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R34
 
 | | Difference-refined structure of fatty acid photodecarboxylase 900 ps following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
