1G7D
 
 | | NMR STRUCTURE OF ERP29 C-DOMAIN | | Descriptor: | ENDOPLASMIC RETICULUM PROTEIN ERP29 | | Authors: | Liepinsh, E, Mkrtchian, S, Barishev, M, Sharipo, A, Ingelman-Sundberg, M, Otting, G. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Thioredoxin fold as homodimerization module in the putative chaperone ERp29: NMR structures of the domains and experimental model of the 51 kDa dimer.
Structure, 9, 2001
|
|
1EJA
 
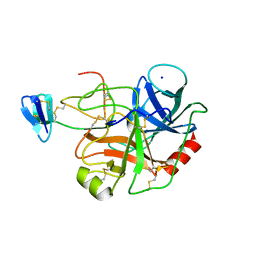 | | STRUCTURE OF PORCINE TRYPSIN COMPLEXED WITH BDELLASTASIN, AN ANTISTASIN-TYPE INHIBITOR | | Descriptor: | BDELLASTASIN, SODIUM ION, TRYPSIN | | Authors: | Rester, U, Moser, M, Huber, R, Bode, W. | | Deposit date: | 2000-03-02 | | Release date: | 2001-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | L-Isoaspartate 115 of porcine beta-trypsin promotes crystallization of its complex with bdellastasin.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
8ARY
 
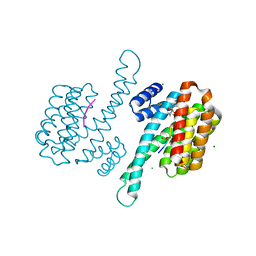 | | Small molecular stabilizer for ERalpha and 14-3-3 (1080273) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-[1-(4-chloranylphenoxy)cyclohexyl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8ZEQ
 
 | |
6ARY
 
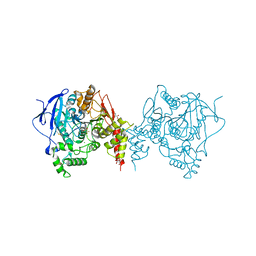 | | Crystal structure of an insecticide-resistant acetylcholinesterase mutant from the malaria vector Anopheles gambiae in complex with a difluoromethyl ketone inhibitor | | Descriptor: | (1S)-2,2-difluoro-1-[1-(pentan-3-yl)-1H-pyrazol-4-yl]ethan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Cheung, J, Mahmood, A, Kalathur, R, Lixuan, L, Carlier, P.R. | | Deposit date: | 2017-08-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.257 Å) | | Cite: | Structure of the G119S Mutant Acetylcholinesterase of the Malaria Vector Anopheles gambiae Reveals Basis of Insecticide Resistance.
Structure, 26, 2018
|
|
9ARY
 
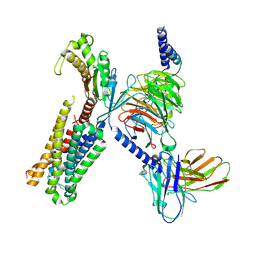 | | Global reconstruction 5-HT2AR bound to 5-HT in complex with a mini-Gq protein and scFv16 obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | 5-hydroxytryptamine receptor 2A, G subunit q (Gi2-mini-Gq chimeric), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Gumpper, R.H, Fay, J.F, Roth, B.L. | | Deposit date: | 2024-02-24 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | The structural diversity of psychedelic drug actions revealed.
Nat Commun, 16, 2025
|
|
2ARY
 
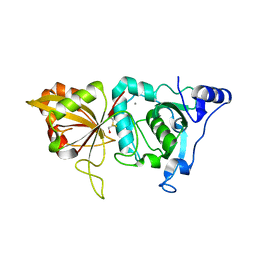 | | Catalytic domain of Human Calpain-1 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, Calpain-1 catalytic subunit | | Authors: | Walker, J.R, Davis, T, Lunin, V, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-22 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structures of Human Calpains 1 and 9 Imply Diverse Mechanisms of Action and Auto-inhibition
J.Mol.Biol., 366, 2007
|
|
7ARY
 
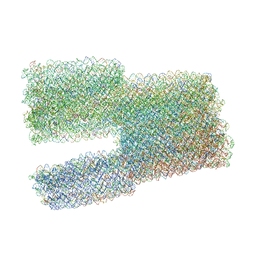 | | Twist-Tower_twist-corrected-variant | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
8WBY
 
 | | Cryo-EM structure of ACE2-B0AT1 complex with JX98 | | Descriptor: | 2-(4-chloranyl-3,5-dimethyl-phenoxy)-~{N}-propan-2-yl-ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R, Hu, Z, Dai, L. | | Deposit date: | 2023-09-10 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Molecular basis of inhibition of the amino acid transporter B 0 AT1 (SLC6A19).
Nat Commun, 15, 2024
|
|
8WBZ
 
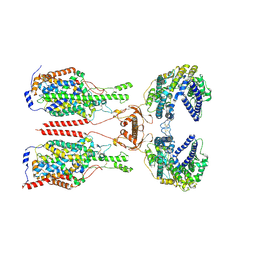 | | Cryo-EM structure of ACE2-B0AT1 complex with JX225 | | Descriptor: | 2-(4-bromanyl-3-methyl-phenoxy)-~{N}-propyl-ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R, Hu, Z, Dai, L. | | Deposit date: | 2023-09-10 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of inhibition of the amino acid transporter B 0 AT1 (SLC6A19).
Nat Commun, 15, 2024
|
|
4ARY
 
 | | Lepidopteran-specific toxin Cry1Ac in complex with receptor specificity determinant GalNAc | | Descriptor: | 1,3-DIAMINOPROPANE, 2-acetamido-2-deoxy-beta-D-galactopyranose, PESTICIDAL CRYSTAL PROTEIN CRY1AC | | Authors: | Derbyshire, D.J, Carroll, J, Ellar, D.J, Li, J. | | Deposit date: | 2012-04-27 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis of Galnac-Dependent Receptor Recognition by B. Thuringiensis Toxin Cry1Ac
To be Published
|
|
4ARX
 
 | | Lepidoptera-specific toxin Cry1Ac from Bacillus thuringiensis ssp. kurstaki HD-73 | | Descriptor: | 1,3-DIAMINOPROPANE, GLYCEROL, PESTICIDAL CRYSTAL PROTEIN CRY1AC | | Authors: | Derbyshire, D.J, Carroll, J, Ellar, D.J, Li, J. | | Deposit date: | 2012-04-27 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Galnac-Dependent Receptor Recognition by B. Thuringiensis Toxin Cry1Ac
To be Published
|
|
3ECS
 
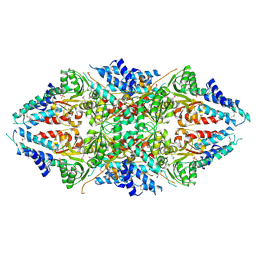 | | Crystal structure of human eIF2B alpha | | Descriptor: | CHLORIDE ION, SULFATE ION, Translation initiation factor eIF-2B subunit alpha | | Authors: | Hiyama, T.B, Ito, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-09-01 | | Release date: | 2009-09-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the alpha subunit of human translation initiation factor 2B
J.Mol.Biol., 392, 2009
|
|
1KMR
 
 | |
4V69
 
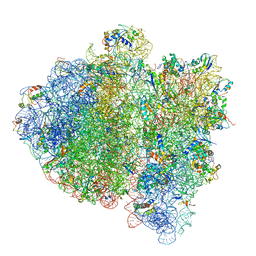 | | Ternary complex-bound E.coli 70S ribosome. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Villa, E, Sengupta, J, Trabuco, L.G, LeBarron, J, Baxter, W.T, Shaikh, T.R, Grassucci, R.A, Nissen, P, Ehrenberg, M, Schulten, K, Frank, J. | | Deposit date: | 2008-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8RCK
 
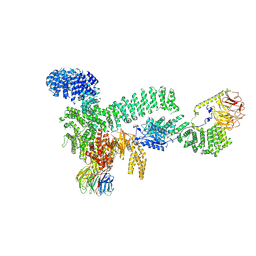 | | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, Focused on one protomer copy. | | Descriptor: | Eukaryotic translation initiation factor 4E-binding protein 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Anandapadamanaban, M, Hay, I.M, Perisic, O, Williams, R.L. | | Deposit date: | 2023-12-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, Focused on one protomer copy.
To Be Published
|
|
8RCH
 
 | | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, overall refinement | | Descriptor: | Eukaryotic translation initiation factor 4E-binding protein 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Anandapadamanaban, M, Hay, I.M, Perisic, O, Williams, R.L. | | Deposit date: | 2023-12-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | cryoEM structure of mTORC1 with the cancer-associated 1455-EWED-1458 duplication variant, overall refinement
To Be Published
|
|
8RCN
 
 | | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, Focused region of mTOR and RAPTOR on one protomer copy. | | Descriptor: | Eukaryotic translation initiation factor 4E-binding protein 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Anandapadamanaban, M, Hay, I.M, Perisic, O, Williams, R.L. | | Deposit date: | 2023-12-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, Focused region of mTOR and RAPTOR on one protomer copy.
To Be Published
|
|
5APO
 
 | | Structure of the yeast 60S ribosomal subunit in complex with Arx1, Alb1 and C-terminally tagged Rei1 | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Greber, B.J, Gerhardy, S, Leitner, A, Leibundgut, M, Salem, M, Boehringer, D, Leulliot, N, Aebersold, R, Panse, V.G, Ban, N. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Insertion of the Biogenesis Factor Rei1 Probes the Ribosomal Tunnel during 60S Maturation.
Cell(Cambridge,Mass.), 164, 2016
|
|
5APN
 
 | | Structure of the yeast 60S ribosomal subunit in complex with Arx1, Alb1 and N-terminally tagged Rei1 | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Greber, B.J, Gerhardy, S, Leitner, A, Leibundgut, M, Salem, M, Boehringer, D, Leulliot, N, Aebersold, R, Panse, V.G, Ban, V. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Insertion of the Biogenesis Factor Rei1 Probes the Ribosomal Tunnel during 60S Maturation.
Cell, 164, 2015
|
|
4V8T
 
 | | Cryo-EM Structure of the 60S Ribosomal Subunit in Complex with Arx1 and Rei1 | | Descriptor: | 25S RIBOSOMAL RNA, 5.8S RIBOSOMAL RNA, 5S RIBOSOMAL RNA, ... | | Authors: | Greber, B.J, Boehringer, D, Montellese, C, Ban, N. | | Deposit date: | 2012-08-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Cryo-Em Structures of Arx1 and Maturation Factors Rei1 and Jjj1 Bound to the 60S Ribosomal Subunit
Nat.Struct.Mol.Biol., 19, 2012
|
|
5XLN
 
 | |
8QZZ
 
 | | Crystal structure of human eIF2 alpha-gamma complexed with PPP1R15A_420-452 | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Yan, Y, Ron, D. | | Deposit date: | 2023-10-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Substrate recruitment via eIF2 gamma enhances catalytic efficiency of a holophosphatase that terminates the integrated stress response.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4UER
 
 | | 40S-eIF1-eIF1A-eIF3-eIF3j translation initiation complex from Lachancea kluyveri | | Descriptor: | 18S RRNA, EIF1, EIF1A, ... | | Authors: | Aylett, C.H.S, Boehringer, D, Erzberger, J.P, Schaefer, T, Ban, N. | | Deposit date: | 2014-12-18 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.47 Å) | | Cite: | Structure of a Yeast 40S-Eif1-Eif1A-Eif3-Eif3J Initiation Complex
Nat.Struct.Mol.Biol., 22, 2015
|
|
5M7W
 
 | | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpG mRNA 5' cap analog (beta-S-ARCA D2) | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(sulfanyl)phosphoryl] hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpG mRNA 5' cap analog (beta-S-ARCA D2)
To Be Published
|
|
