6TEL
 
 | | Crystal structure of Dot1L in complex with an inhibitor (compound 10). | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, POTASSIUM ION, ... | | Authors: | Scheufler, C, Stauffer, F, Be, C, Moebitz, H. | | Deposit date: | 2019-11-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | New Potent DOT1L Inhibitors forin VivoEvaluation in Mouse.
Acs Med.Chem.Lett., 10, 2019
|
|
6TEN
 
 | | Crystal structure of Dot1L in complex with an inhibitor (compound 11). | | Descriptor: | 3-[(4-azanyl-6-methoxy-1,3,5-triazin-2-yl)amino]-4-[[(~{S})-[2,2-bis(fluoranyl)-1,3-benzodioxol-4-yl]-(3-chloranylpyridin-2-yl)methyl]amino]benzenesulfonamide, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Scheufler, C, Stauffer, F, Be, C, Moebitz, H. | | Deposit date: | 2019-11-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | New Potent DOT1L Inhibitors forin VivoEvaluation in Mouse.
Acs Med.Chem.Lett., 10, 2019
|
|
8SVF
 
 | | BAP1/ASXL1 bound to the H2AK119Ub Nucleosome | | Descriptor: | DNA/RNA (187-MER), DNA/RNA (327-MER), Histone H2A type 1, ... | | Authors: | Thomas, J.F, Valencia-Sanchez, M.I, Armache, K.-J. | | Deposit date: | 2023-05-16 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of histone H2A lysine 119 deubiquitination by Polycomb repressive deubiquitinase BAP1/ASXL1.
Sci Adv, 9, 2023
|
|
6FML
 
 | | CryoEM Structure INO80core Nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin related protein 5, ... | | Authors: | Eustermann, S, Schall, K, Kostrewa, D, Strauss, M, Hopfner, K. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Structural basis for ATP-dependent chromatin remodelling by the INO80 complex.
Nature, 556, 2018
|
|
8SKZ
 
 | |
8THU
 
 | |
8T9F
 
 | |
8T9H
 
 | |
8T3W
 
 | | Structure of Bre1-nucleosome complex - state2 | | Descriptor: | 601 DNA strand 1, 601 DNA strand 2, E3 ubiquitin-protein ligase BRE1, ... | | Authors: | Zhao, F, Hicks, C.W, Wolberger, C. | | Deposit date: | 2023-06-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Mechanism of histone H2B monoubiquitination by Bre1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T3Y
 
 | | Structure of Bre1-nucleosome complex - state1 | | Descriptor: | 601 DNA Strand 1, 601 DNA Strand 2, E3 ubiquitin-protein ligase BRE1, ... | | Authors: | Zhao, F, Hicks, C.W, Wolberger, C. | | Deposit date: | 2023-06-08 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Mechanism of histone H2B monoubiquitination by Bre1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T3T
 
 | | Structure of Bre1-nucleosome complex - state3 | | Descriptor: | 601 DNA strand 1, 601 DNA strand 2, E3 ubiquitin-protein ligase BRE1, ... | | Authors: | Zhao, F, Hicks, C.W, Wolberger, C. | | Deposit date: | 2023-06-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Mechanism of histone H2B monoubiquitination by Bre1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6D07
 
 | |
6W5I
 
 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class01) | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6W5N
 
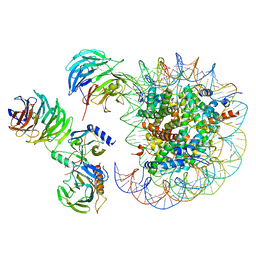 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class05) | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6W5M
 
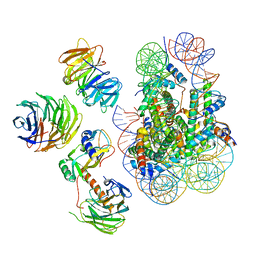 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class02) | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
3X1T
 
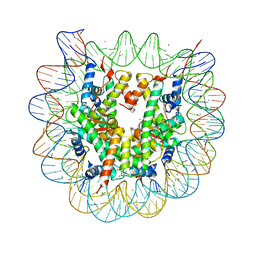 | |
2CV5
 
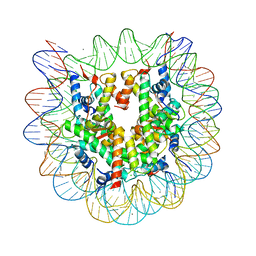 | | Crystal structure of human nucleosome core particle | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.a, ... | | Authors: | Tsunaka, Y, Kajimura, N, Tate, S, Morikawa, K. | | Deposit date: | 2005-05-31 | | Release date: | 2005-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle
Nucleic Acids Res., 33, 2005
|
|
9MVY
 
 | | Crystal structure of ZMET2 in complex with unmethylated CTG DNA and a histone H3Kc9me2 peptide | | Descriptor: | C49 ssDNA, DNA (cytosine-5)-methyltransferase 1, Histone H3.2, ... | | Authors: | Herle, G, Fang, J, Song, J. | | Deposit date: | 2025-01-16 | | Release date: | 2025-05-21 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of an Unfavorable de Novo DNA Methylation Complex of Plant Methyltransferase ZMET2.
J.Mol.Biol., 437, 2025
|
|
9N6K
 
 | | 2.88 A S.cerevisiae Chd1[L886G/L889G/L891G]-nucleosome 2:1 complex with DNA-binding domain | | Descriptor: | Chromo domain-containing protein 1, DNA Lagging Strand, DNA Tracking Strand, ... | | Authors: | Nodelman, I.M, Folkwein, H.J, Armache, J.-P, Bowman, G.D. | | Deposit date: | 2025-02-05 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A competitive regulatory mechanism of the Chd1 remodeler is integral to distorting nucleosomal DNA.
Nat.Struct.Mol.Biol., 2025
|
|
9N6H
 
 | | 2.54 A S.cerevisiae Chd1[L886G/L889G/L891G]-nucleosome 1:1 complex | | Descriptor: | Chromo domain-containing protein 1, DNA Lagging Strand, DNA Tracking Strand, ... | | Authors: | Nodelman, I.M, Folkwein, H.J, Armache, J.-P, Bowman, G.D. | | Deposit date: | 2025-02-05 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | A competitive regulatory mechanism of the Chd1 remodeler is integral to distorting nucleosomal DNA.
Nat.Struct.Mol.Biol., 2025
|
|
9N6I
 
 | | 2.61 A S.cerevisiae Chd1[L886G/L889G/L891G]-nucleosome 2:1 complex | | Descriptor: | Chromo domain-containing protein 1, DNA Lagging Strand, DNA Tracking Strand, ... | | Authors: | Nodelman, I.M, Folkwein, H.J, Armache, J.-P, Bowman, G.D. | | Deposit date: | 2025-02-05 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | A competitive regulatory mechanism of the Chd1 remodeler is integral to distorting nucleosomal DNA.
Nat.Struct.Mol.Biol., 2025
|
|
8G6Q
 
 | | H2AK119ub-modified nucleosome ubiquitin position 1 | | Descriptor: | 601 DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Hicks, C.W, Wolberger, C, Keogh, M. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-21 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Ubiquitinated histone H2B as gatekeeper of the nucleosome acidic patch.
Nucleic Acids Res., 52, 2024
|
|
8G6S
 
 | | H2AK119ub-modified nucleosome ubiquitin position 2 | | Descriptor: | 601 DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Hicks, C.W, Wolberger, C, Keogh, M. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Ubiquitinated histone H2B as gatekeeper of the nucleosome acidic patch.
Nucleic Acids Res., 52, 2024
|
|
8X2Z
 
 | | The class2 of piccolo NuA4 bound to the H2A.Z nucleosome complex at harboring state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X32
 
 | | The piccolo NuA4 bound to the H2A.Z nucleosome-H4KQ Complex with Ac-CoA at resetting state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
