5KYO
 
 | | Crystal Structure of CYP101J2 | | Descriptor: | CYP101J2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Unterweger, B, Drinkwater, N, Dumsday, G.J, McGowan, S. | | Deposit date: | 2016-07-22 | | Release date: | 2017-01-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of cytochrome P450 monooxygenase CYP101J2 from Sphingobium yanoikuyae strain B2.
Proteins, 85, 2017
|
|
8FGH
 
 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(5-(2-(dimethylamino)ethyl)-2,3-difluorophenethyl)-4-methoxypyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[2-(dimethylamino)ethyl]-2,3-difluorophenyl}ethyl)-4-methoxypyridin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
6WAB
 
 | | Crystal structure of human galectin-4 C-terminal carbohydrate recognition domain in complex with galactose derivative | | Descriptor: | 2,6-anhydro-1,4-dideoxy-1-[4-(4-fluorophenyl)-1H-1,2,3-triazol-1-yl]-4-(4-phenyl-1H-1,2,3-triazol-1-yl)-D-glycero-L-manno-heptitol, GLYCEROL, Galectin-4 | | Authors: | Go, R.M, Kishor, C, Blanchard, H. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of human galectin-4 C-terminal carbohydrate recognition domain in complex with galactose derivative
To Be Published
|
|
6WB4
 
 | | Microbiome-derived Acarbose Kinase Mak1 Labeled with selenomethionine | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Acarbose Kinase Mak1, ... | | Authors: | Jeffrey, P.D, Balaich, J.N, Estrella, M.A, Donia, M.S. | | Deposit date: | 2020-03-26 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | The human microbiome encodes resistance to the antidiabetic drug acarbose.
Nature, 600, 2021
|
|
5A3A
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes (Apo form) | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, SIR2 FAMILY PROTEIN, ... | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
6WB5
 
 | | Microbiome-derived Acarbose Kinase Mak1 as a Complex with Acarbose and AMP-PNP | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Acarbose Kinase Mak1, MANGANESE (II) ION, ... | | Authors: | Jeffrey, P.D, Balaich, J.N, Estrella, M.A, Donia, M.S. | | Deposit date: | 2020-03-26 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | The human microbiome encodes resistance to the antidiabetic drug acarbose.
Nature, 600, 2021
|
|
8FGG
 
 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(5-(2-(dimethylamino)ethyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[2-(dimethylamino)ethyl]-2,3-difluorophenyl}ethyl)-4-methylpyridin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
5AF1
 
 | |
8EVE
 
 | | HUMAN DNA POLYMERASE ETA INSERTION COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2022-10-20 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The peroxidation-derived DNA adduct, 6-oxo-M 1 dG, is a strong block to replication by human DNA polymerase eta.
J.Biol.Chem., 299, 2023
|
|
6WB7
 
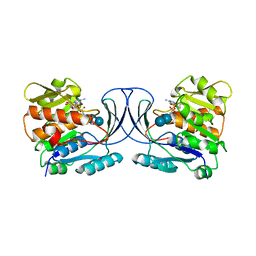 | | Acarbose Kinase AcbK as a Complex with Acarbose and AMP-PNP | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Acarbose 7(IV)-phosphotransferase, MANGANESE (II) ION, ... | | Authors: | Jeffrey, P.D, Balaich, J.N, Estrella, M.A, Donia, M.S. | | Deposit date: | 2020-03-26 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The human microbiome encodes resistance to the antidiabetic drug acarbose.
Nature, 600, 2021
|
|
5ADN
 
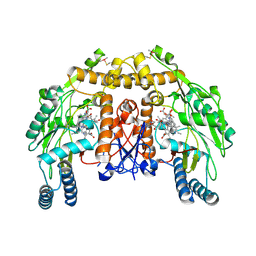 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 7-(((3-((Dimethylamino)methyl)phenyl)amino)methyl) quinolin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[[3-[(dimethylamino)methyl]phenyl]amino]methyl]quinolin-2-amine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-08-20 | | Release date: | 2015-10-28 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phenyl Ether- and Aniline-Containing 2-Aminoquinolines as Potent and Selective Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
5L8F
 
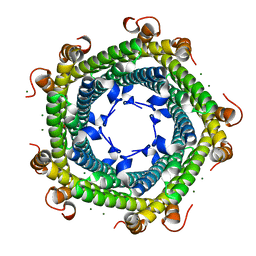 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E32A, E62A, H65A. | | Descriptor: | MAGNESIUM ION, Rru_A0973, TRIETHYLENE GLYCOL | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E32A, E62A, H65A.
To be published
|
|
5L92
 
 | |
8HS7
 
 | |
5L9N
 
 | | Structure of uridylylated GlnB from Escherichia coli bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Nitrogen regulatory protein P-II 1 | | Authors: | Rubio, V, Palanca, C. | | Deposit date: | 2016-06-10 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Effects of T-loop modification on the PII-signalling protein: structure of uridylylated Escherichia coli GlnB bound to ATP.
Environ Microbiol Rep, 9, 2017
|
|
5L4I
 
 | | Crystal Structure of Human Transthyretin in Complex with Clonixin | | Descriptor: | 2-(3-chloro-2-methylanilino)pyridine-3-carboxylic acid, SODIUM ION, Transthyretin | | Authors: | Grundstrom, C, Hall, M, Zhang, J, Olofsson, A, Andersson, P, Sauer-Eriksson, A.E. | | Deposit date: | 2016-05-25 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Based Virtual Screening Protocol for in Silico Identification of Potential Thyroid Disrupting Chemicals Targeting Transthyretin.
Environ. Sci. Technol., 50, 2016
|
|
5LBG
 
 | | Crystal structure of the Mycobacterium tuberculosis L,D-transpeptidase-2 (LdtMt2) BC-module with faropenem-derived adduct at the active site cysteine-354 | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, L,D-transpeptidase 2 | | Authors: | Steiner, E.M, Schnell, R, Schneider, G. | | Deposit date: | 2016-06-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Binding and processing of beta-lactam antibiotics by the transpeptidase LdtMt2 from Mycobacterium tuberculosis.
FEBS J., 284, 2017
|
|
8HRV
 
 | | dutpase of helicobacter pylori 26695 | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Kumari, K, Gourinath, S. | | Deposit date: | 2022-12-16 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | dutpase of helicobacter pylori 26695
To Be Published
|
|
5ACU
 
 | | VIM-2-NAT, Discovery of novel inhibitor scaffolds against the metallo- beta-lactamase VIM-2 by SPR based fragment screening | | Descriptor: | BETA-LACTAMASE, CHLORIDE ION, HYDROXIDE ION, ... | | Authors: | Christopeit, T, Carlsen, T.J.O, Helland, R, Leiros, H.K.S. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Novel Inhibitor Scaffolds Against the Metallo-Beta-Lactamase Vim-2 by Spr Based Fragment Screening
J.Med.Chem., 58, 2015
|
|
5AD2
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | (3R)-4-(2-{4-[1-(3-chloro[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidinyl]phenoxy}ethyl)-1,3-dimethyl-2-piperazinone, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
6VLI
 
 | | Crystal structure of transcriptional regulator from bacteriophage 186 | | Descriptor: | ACETATE ION, ACETIC ACID, Regulatory protein CII, ... | | Authors: | Truong, J.Q, Panjikar, S, Bruning, J.B, Shearwin, K.E. | | Deposit date: | 2020-01-24 | | Release date: | 2021-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.100021 Å) | | Cite: | Crystal structure of a transcriptional regulator from bacteriophage 186
To Be Published
|
|
6VMH
 
 | |
6VPE
 
 | |
8EO5
 
 | | Crystal structure of the class A beta-lactamase precursor LRA-5 from an Alaskan soil metagenome at 1.8 Angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LRA-5 | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
8EV4
 
 | | NlpC B3 - Trichomonas Vaginalis | | Descriptor: | Clan CA, family C40, NlpC/P60 superfamily cysteine peptidase | | Authors: | Barnett, M.J, Goldstone, D.C. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | NlpC/P60 peptidoglycan hydrolases of Trichomonas vaginalis have complementary activities that empower the protozoan to control host-protective lactobacilli.
Plos Pathog., 19, 2023
|
|
