7ZOC
 
 | |
5ZJY
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, LYS-LYS-ARG-TYR-SER-ARG-2JN-GLN-LEU-LEU-2JN-PHE | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, B, Uhring, M. | | Deposit date: | 2018-03-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
5ZK5
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, LYS-ARG-TYR-SER-ARG-GLU-GLN-LEU-LEU-MK8-PHE-GLN-ARG-MK8 | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, F, Uhring, M. | | Deposit date: | 2018-03-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
5G01
 
 | | An unusual natural product primary sulfonamide: synthesis, carbonic anhydrase inhibition and protein x-ray structure of Psammaplin C | | Descriptor: | CARBONIC ANHYDRASE 2, PSAMMAPLIN C, SODIUM ION, ... | | Authors: | Mujumdar, P, Supuran, C.T, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Unusual Natural Product Primary Sulfonamide: Synthesis, Carbonic Anhydrase Inhibition and Protein X-Ray Structures of Psammaplin C.
J.Med.Chem., 59, 2016
|
|
5ZK9
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, ACE-ARG-ILE-ILE-TYR-SER-ARG-MK8-GLN-LEU-LEU-MK8-LEU-LYS-NH2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, F, Uhring, M. | | Deposit date: | 2018-03-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
5ZK7
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, ACE-ARG-TYR-SER-ARG-MK8-GLN-LEU-LEU-MK8-LEU-PHE-ARG-NH2, CHLORIDE ION, ... | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, F, Uhring, M. | | Deposit date: | 2018-03-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
5ZML
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 1,2-ETHANEDIOL, ACE-LYS-LYS-ARG-TYR-SER-ARG-MK8-GLN-LEU-LEU-MK8-PHE-ARG-ARG, Eukaryotic translation initiation factor 4E, ... | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Yano, J. | | Deposit date: | 2018-04-04 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
2HRK
 
 | |
2HSM
 
 | |
2HSN
 
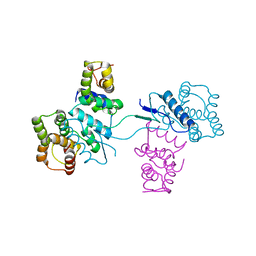 | | Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes | | Descriptor: | GU4 nucleic-binding protein 1, Methionyl-tRNA synthetase, cytoplasmic | | Authors: | Simader, H, Koehler, C, Basquin, J, Suck, D. | | Deposit date: | 2006-07-22 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes.
Nucleic Acids Res., 34, 2006
|
|
5OMF
 
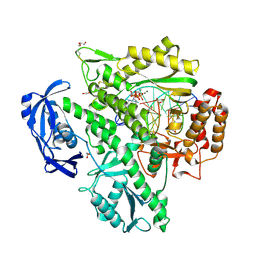 | | Closed, ternary structure of KOD DNA polymerase | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*CP*AP*C)-3'), ... | | Authors: | Kropp, H.M, Betz, K, Wirth, J, Diederichs, K, Marx, A. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Crystal structures of ternary complexes of archaeal B-family DNA polymerases.
PLoS ONE, 12, 2017
|
|
6XB7
 
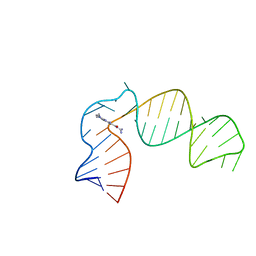 | | IRES-targeting Small Molecule Inhibits Enterovirus 71 Replication via Allosteric Stabilization of a Ternary Complex | | Descriptor: | 3-amino-N-(diaminomethylidene)-5-(dimethylamino)-6-(phenylethynyl)pyrazine-2-carboxamide, RNA (41-MER) | | Authors: | Tolbert, B.S, Davila, J, Sugarman, A.L, Chiu, L.Y. | | Deposit date: | 2020-06-05 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | IRES-targeting small molecule inhibits enterovirus 71 replication via allosteric stabilization of a ternary complex.
Nat Commun, 11, 2020
|
|
7PGB
 
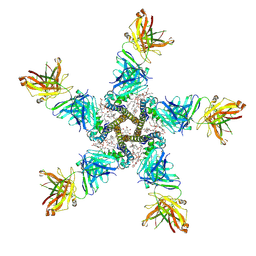 | | NaV_Ae1/Sp1CTD_pore-SAT09 complex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-NITROBENZOIC ACID, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGF
 
 | |
7PGG
 
 | | NaVAb1p detergent (DM) | | Descriptor: | 2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}ethyl heptadecanoate, Ion transport protein | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4CBC
 
 | |
1K6O
 
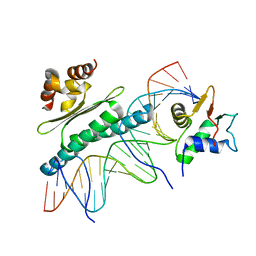 | | Crystal Structure of a Ternary SAP-1/SRF/c-fos SRE DNA Complex | | Descriptor: | 5'-D(*CP*AP*CP*AP*GP*GP*AP*TP*GP*TP*CP*CP*AP*TP*AP*TP*TP*AP*GP*GP*AP*CP*A)-3', 5'-D(*TP*GP*TP*CP*CP*TP*AP*AP*TP*AP*TP*GP*GP*AP*CP*AP*TP*CP*CP*TP*GP*TP*G)-3', ETS-domain protein ELK-4, ... | | Authors: | Mo, Y, Ho, W, Johnston, K, Marmorstein, R. | | Deposit date: | 2001-10-16 | | Release date: | 2002-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex.
J.Mol.Biol., 314, 2001
|
|
3D2V
 
 | |
4KXT
 
 | |
1LRT
 
 | | CRYSTAL STRUCTURE OF TERNARY COMPLEX OF TRITRICHOMONAS FOETUS INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE: STRUCTURAL CHARACTERIZATION OF NAD+ SITE IN MICROBIAL ENZYME | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, ... | | Authors: | Gan, L, Petsko, G.A, Hedstrom, L. | | Deposit date: | 2002-05-15 | | Release date: | 2003-07-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a ternary complex of Tritrichomonas foetus inosine
5'-monophosphate dehydrogenase: NAD+ orients the active site loop for catalysis
Biochemistry, 41, 2003
|
|
1KPD
 
 | |
3V1U
 
 | | Crystal structure of a beta-ketoacyl reductase FabG4 from Mycobacterium tuberculosis H37Rv complexed with NAD+ and Hexanoyl-CoA at 2.5 Angstrom resolution | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, 3-oxoacyl-(Acyl-carrier-protein) reductase, HEXANOYL-COENZYME A, ... | | Authors: | Dutta, D, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2011-12-10 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hexanoyl-CoA bound to beta-ketoacyl reductase FabG4 of Mycobacterium tuberculosis
Biochem.J., 450, 2013
|
|
1RQ4
 
 | | Crystallographic Analysis of the Interaction of Nitric Oxide with Quaternary-T Human Hemoglobin, HEMOGLOBIN EXPOSED TO NO UNDER AEROBIC CONDITIONS | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, NITRIC OXIDE, ... | | Authors: | Chan, N.L, Kavanaugh, J.S, Rogers, P.H, Arnone, A. | | Deposit date: | 2003-12-04 | | Release date: | 2003-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystallographic analysis of the interaction of nitric oxide with quaternary-T human hemoglobin.
Biochemistry, 43, 2004
|
|
5G0W
 
 | | InhA in complex with a DNA encoded library hit | | Descriptor: | (2S,4S)-N-methyl-4-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-[[4-(pyrazol-1-ylmethyl)phenyl]carbonylamino]butanoyl]amino]-1-(phenylcarbonyl)pyrrolidine-2-carboxamide, ENOYL-ACYL CARRIER PROTEIN REDUCTASE, MAGNESIUM ION, ... | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Cofactor-Specific, Bactericidal Mycobacterium Tuberculosis Inha Inhibitors Using DNA-Encoded Library Technology
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1RQ3
 
 | | Crystallographic Analysis of the Interaction of Nitric Oxide with Quaternary-T Human Deoxyhemoglobin, Deoxyhemoglobin | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chan, N.L, Kavanaugh, J.S, Rogers, P.H, Arnone, A. | | Deposit date: | 2003-12-04 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystallographic analysis of the interaction of nitric oxide with quaternary-T human hemoglobin.
Biochemistry, 43, 2004
|
|
