1XXH
 
 | | ATPgS Bound E. Coli Clamp Loader Complex | | Descriptor: | DNA polymerase III subunit gamma, DNA polymerase III, delta prime subunit, ... | | Authors: | Kazmirski, S.L, Podobnik, M, Weitze, T.F, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2004-11-05 | | Release date: | 2004-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural analysis of the inactive state of the Escherichia coli DNA polymerase clamp-loader complex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XXI
 
 | | ADP Bound E. coli Clamp Loader Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase III subunit gamma, DNA polymerase III, ... | | Authors: | Kazmirski, S.L, Podobnik, M, Weitze, T.F, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2004-11-05 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural analysis of the inactive state of the Escherichia coli DNA polymerase clamp-loader complex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2J5C
 
 | | Rational conversion of substrate and product specificity in a monoterpene synthase. Structural insights into the molecular basis of rapid evolution. | | Descriptor: | 1,8-CINEOLE SYNTHASE, BETA-MERCAPTOETHANOL | | Authors: | Kampranis, S.C, Ioannidis, D, Purvis, A, Mahrez, W, Ninga, E, Katerelos, N.A, Anssour, S, Dunwell, J.M, Makris, A.M, Goodenough, P.W, Johnson, C.B. | | Deposit date: | 2006-09-14 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational Conversion of Substrate and Product Specificity in a Salvia Monoterpene Synthase: Structural Insights Into the Evolution of Terpene Synthase Function.
Plant Cell, 19, 2007
|
|
2Y0Q
 
 | |
2MAO
 
 | |
2L1G
 
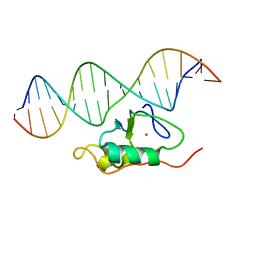 | | RDC refined solution structure of the THAP zinc finger of THAP1 in complex with its 16bp RRM1 DNA target | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*GP*TP*GP*TP*GP*GP*GP*CP*AP*GP*CP*G)-3'), DNA (5'-D(P*CP*GP*CP*TP*GP*CP*CP*CP*AP*CP*AP*CP*AP*AP*GP*C)-3'), THAP domain-containing protein 1, ... | | Authors: | Campagne, S, Gervais, V, Saurel, O, Milon, A. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | RDC refined solution structure of the THAP zinc finger of THAP1 in complex with its 16bp RRM1 DNA target
To be published
|
|
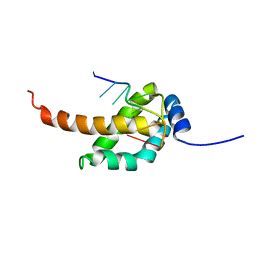
2MAP
 
 | |
2Y0T
 
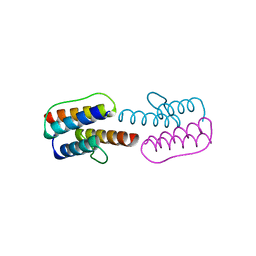 | |
2KO0
 
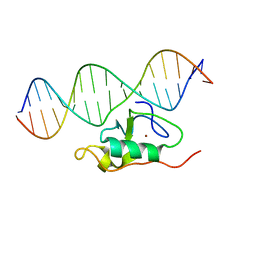 | | Solution structure of the THAP zinc finger of THAP1 in complex with its DNA target | | Descriptor: | RRM1, THAP domain-containing protein 1, ZINC ION | | Authors: | Campagne, S, Gervais, V, Saurel, O, Milon, A. | | Deposit date: | 2009-09-08 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of specific DNA-recognition by the THAP zinc finger
Nucleic Acids Res., 2010
|
|
3WLD
 
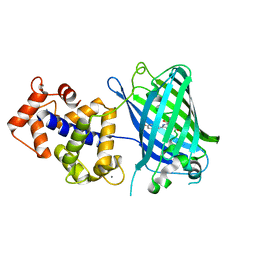 | | Crystal structure of monomeric GCaMP6m | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Ding, J, Luo, A.F, Hu, L.Y, Wang, D.C, Shao, F. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the ultrasensitive calcium indicator GCaMP6.
Sci China Life Sci, 57, 2014
|
|
3NV6
 
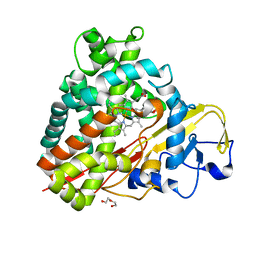 | | Crystal Structure of Camphor-Bound CYP101D2 | | Descriptor: | CAMPHOR, Cytochrome P450, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
2KXW
 
 | |
2MP1
 
 | |
2LFW
 
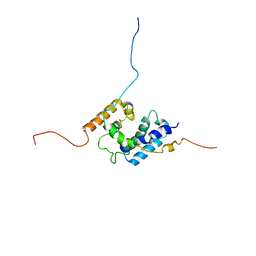 | | NMR structure of the PhyRSL-NepR complex from Sphingomonas sp. Fr1 | | Descriptor: | NepR anti sigma factor, PhyR sigma-like domain | | Authors: | Campagne, S, Damberger, F.F, Vorholt, J.A, Allain, F.H.-T. | | Deposit date: | 2011-07-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sigma factor mimicry in the general stress response of Alphaproteobacteria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3KG2
 
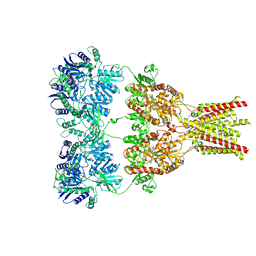 | | AMPA subtype ionotropic glutamate receptor in complex with competitive antagonist ZK 200775 | | Descriptor: | Glutamate receptor 2, beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sobolevsky, A.I, Rosconi, M.P, Gouaux, E. | | Deposit date: | 2009-10-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor.
Nature, 462, 2009
|
|
6XSR
 
 | |
2ZWU
 
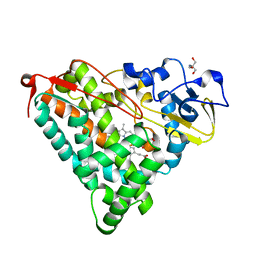 | | Crystal Structure of Camphor Soaked Ferric Cytochrome P450cam | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Camphor 5-monooxygenase, ... | | Authors: | Sakurai, K, Shimada, H, Hayashi, T, Tsukihara, T. | | Deposit date: | 2008-12-18 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Substrate binding induces structural changes in cytochrome P450cam
Acta Crystallogr.,Sect.F, 65, 2009
|
|
9B00
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with berberine analog of chloramphenicol CAM-BER, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.80A resolution | | Descriptor: | 13-(2-{[(1R,2R)-1,3-dihydroxy-1-(4-nitrophenyl)propan-2-yl]amino}-2-oxoethyl)-9,10-dimethoxy-5,6-dihydro-2H-[1,3]dioxolo[4,5-g]isoquinolino[3,2-a]isoquinolin-7-ium, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Batool, Z, Pavlova, J.A, Paranjpe, M.N, Tereshchenkov, A.G, Lukianov, D.A, Osterman, I.A, Bogdanov, A.A, Sumbatyan, N.V, Polikanov, Y.S. | | Deposit date: | 2024-03-11 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Berberine analog of chloramphenicol exhibits a distinct mode of action and unveils ribosome plasticity.
Structure, 32, 2024
|
|
3WLC
 
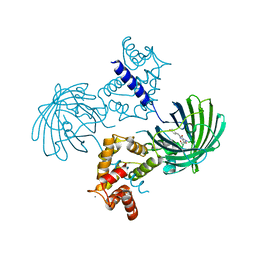 | | Crystal structure of dimeric GCaMP6m | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Ding, J, Luo, A.F, Hu, L.Y, Wang, D.C, Shao, F. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of the ultrasensitive calcium indicator GCaMP6.
Sci China Life Sci, 57, 2014
|
|
4E07
 
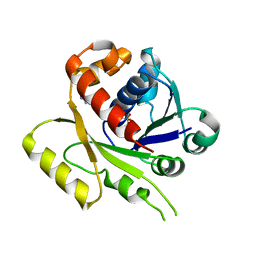 | | ParF-AMPPCP-C2221 form | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Plasmid partitioning protein ParF | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4F9K
 
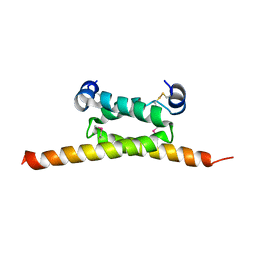 | | Crystal Structure of human cAMP-dependent protein kinase type I-beta regulatory subunit (fragment 11-73), Northeast Structural Genomics Consortium (NESG) Target HR8613A | | Descriptor: | cAMP-dependent protein kinase type I-beta regulatory subunit | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Shastry, R, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-18 | | Release date: | 2012-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of human cAMP-dependent protein kinase type I-beta regulatory subunit (fragment 11-73), Northeast Structural Genomics Consortium (NESG) Target HR8613A
To be Published
|
|
5VOT
 
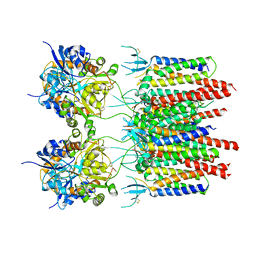 | | Structure of AMPA receptor-TARP complex | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Chen, S, Zhao, Y, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
5VOV
 
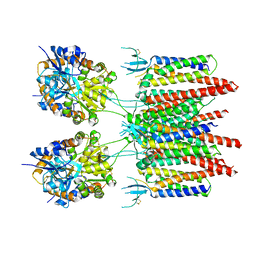 | | Structure of AMPA receptor-TARP complex | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Zhao, Y, Chen, S, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
5VOU
 
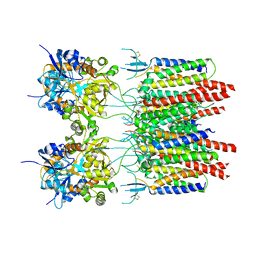 | | Structure of AMPA receptor-TARP complex | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Chen, S, Zhao, Y, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
3U9B
 
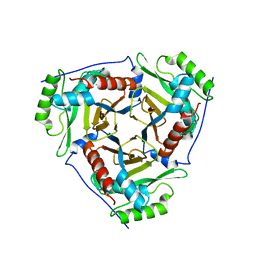 | | Structure of apo-CATI | | Descriptor: | Chloramphenicol acetyltransferase | | Authors: | Biswas, T, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2011-10-18 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis for substrate versatility of chloramphenicol acetyltransferase CAT(I).
Protein Sci., 21, 2012
|
|
