4ISG
 
 | | Human glucokinase in complex with novel activator (2S)-3-cyclohexyl-2-[4-(methylsulfonyl)-2-oxopiperazin-1-yl]-N-(1,3-thiazol-2-yl)propanamide | | Descriptor: | (2S)-3-cyclohexyl-2-[4-(methylsulfonyl)-2-oxopiperazin-1-yl]-N-(1,3-thiazol-2-yl)propanamide, Glucokinase, IODIDE ION, ... | | Authors: | Hosfield, D, Skene, R.J. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Design, synthesis and SAR of novel glucokinase activators.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1EPR
 
 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-135,040 | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide | | Authors: | Badasso, M, Crawford, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1TAE
 
 | |
4J6Q
 
 | |
3E4Q
 
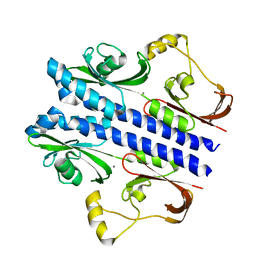 | | Crystal structure of apo DctB | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, CALCIUM ION | | Authors: | Zhou, Y.F, Nan, J, Nan, B.Y, Liang, Y.H, Panjikar, S, Su, X.D. | | Deposit date: | 2008-08-12 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain.
J.Mol.Biol., 383, 2008
|
|
2UXC
 
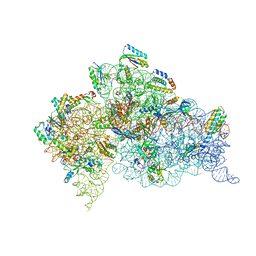 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA UCGU in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON ACGA, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
4NCK
 
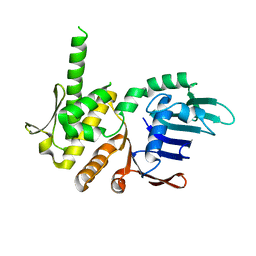 | | Crystal Structure of Pyrococcus furiosis Rad50 R797G mutation | | Descriptor: | CHLORIDE ION, DNA double-strand break repair Rad50 ATPase, MAGNESIUM ION, ... | | Authors: | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
2QI0
 
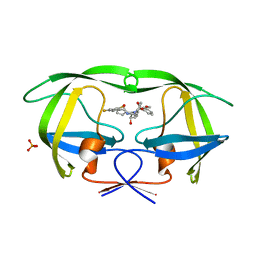 | |
3GB7
 
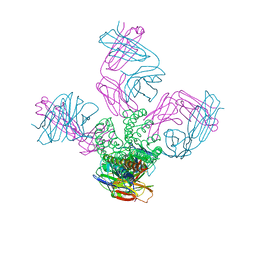 | | Potassium Channel KcsA-Fab complex in Li+ | | Descriptor: | DIACYL GLYCEROL, NICKEL (II) ION, Voltage-gated potassium channel, ... | | Authors: | Thompson, A.N, Ilsoo, K, Panosian, T.D, Iverson, T.M, Allen, T.W, Nimigean, C.M. | | Deposit date: | 2009-02-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Mechanism of potassium-channel selectivity revealed by Na(+) and Li(+) binding sites within the KcsA pore.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3WWR
 
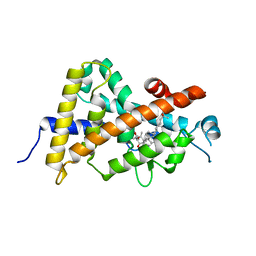 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 1-((((1S,2R,6R,Z)-2,6-dihydroxy-4-((E)-2-((1R,3aS,7aR)-1-((R)-6-hydroxy-6-methylheptan-2-yl)-7a-methylhexahydro-1H-inden-4(2H)-ylidene)ethylidene)-3-methylenecyclohexyl)oxy)methyl)cyclopropanecarbonitrile | | Descriptor: | 1-({[(1R,2S,3R,5Z,7E,14beta,17alpha)-1,3,25-trihydroxy-9,10-secocholesta-5,7,10-trien-2-yl]oxy}methyl)cyclopropanecarbonitrile, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Synthesis and biological activities of vitamin D3 derivatives with cyanoalkyl side chain at C-2 position.
J.Steroid Biochem.Mol.Biol., 148, 2015
|
|
4NCH
 
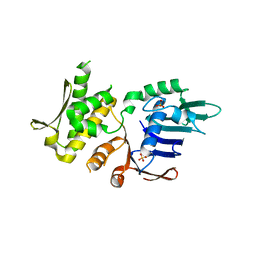 | | Crystal Structure of Pyrococcus furiosis Rad50 L802W mutation | | Descriptor: | DNA double-strand break repair Rad50 ATPase, SULFATE ION | | Authors: | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
4JJO
 
 | |
4JL4
 
 | |
4B4Q
 
 | | Crystal Structure of the lectin domain of F18 fimbrial adhesin FedF in complex with blood group A type 1 hexasaccharide | | Descriptor: | F18 FIMBRIAL ADHESIN AC, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Moonens, K, Bouckaert, J, Coddens, A, Tran, T, Panjikar, S, De Kerpel, M, Cox, E, Remaut, H, De Greve, H. | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight in Histo-Blood Group Binding by the F18 Fimbrial Adhesin Fedf.
Mol.Microbiol., 86, 2012
|
|
4B4R
 
 | | Crystal Structure of the lectin domain of F18 fimbrial adhesin FedF in complex with blood group B type 1 hexasaccharide | | Descriptor: | F18 FIMBRIAL ADHESIN AC, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Moonens, K, Bouckaert, J, Coddens, A, Tran, T, Panjikar, S, De Kerpel, M, Cox, E, Remaut, H, De Greve, H. | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insight in Histo-Blood Group Binding by the F18 Fimbrial Adhesin Fedf.
Mol.Microbiol., 86, 2012
|
|
4ATL
 
 | | Crystal structure of Raucaffricine glucosidase in complex with Glucose | | Descriptor: | RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, beta-D-glucopyranose | | Authors: | Xia, L, Rajendran, C, Ruppert, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2012-05-08 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | High Speed X-Ray Analysis of Plant Enzymes at Room Temperature
Phytochemistry, 91, 2013
|
|
4ATD
 
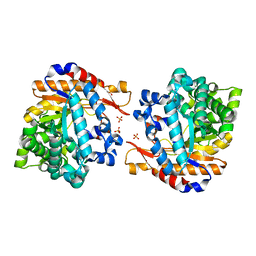 | | Crystal structure of native Raucaffricine glucosidase | | Descriptor: | RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, SULFATE ION | | Authors: | Xia, L, Rajendran, C, Ruppert, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2012-05-05 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High Speed X-Ray Analysis of Plant Enzymes at Room Temperature
Phytochemistry, 91, 2013
|
|
3FDQ
 
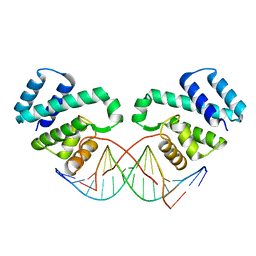 | |
3EY7
 
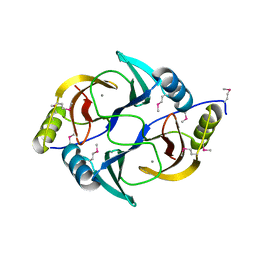 | | Structure from the mobile metagenome of V. Cholerae. Integron cassette protein VCH_CASS1 | | Descriptor: | Biphenyl-2,3-diol 1,2-dioxygenase III-related protein, CALCIUM ION | | Authors: | Harrop, S.J, Deshpande, C.N, Sureshan, V, Boucher, Y, Xu, X, Cui, H, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-20 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure from the mobile metagenome of V. Cholerae. Integron cassette protein VCH_CASS1
To be Published
|
|
5RWT
 
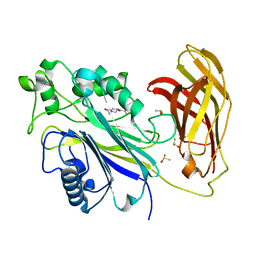 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z32327641 | | Descriptor: | DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1, [4-(cyclopropanecarbonyl)piperazin-1-yl](furan-2-yl)methanone | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
3GHJ
 
 | | Crystal structure from the mobile metagenome of Halifax Harbour Sewage Outfall: Integron Cassette Protein HFX_CASS4 | | Descriptor: | Putative integron gene cassette protein | | Authors: | Sureshan, V, Deshpande, C, Harrop, S.J, Kudritska, M, Koenig, J.E, Evdokimova, E, Kim, Y, Edwards, A.M, Savchenko, A, Joachimiak, A, Doolittle, W.F, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-03 | | Release date: | 2009-03-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Structure from the mobile metagenome of Halifax Harbour Sewage Outfall: Integron Cassette Protein HFX_CASS4
To be Published
|
|
4JB9
 
 | | Crystal structure of antibody VRC06 in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, antibody VRC06 heavy chain, antibody VRC06 light chain, ... | | Authors: | Kwon, Y.D, Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2013-02-19 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization.
Science, 340, 2013
|
|
2OOL
 
 | | Crystal structure of the chromophore-binding domain of an unusual bacteriophytochrome RpBphP3 from R. palustris | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Sensor protein | | Authors: | Yang, X, Stojkovic, E.A, Kuk, J, Moffat, K. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QI7
 
 | | Crystal structure of protease inhibitor, MIT-2-AD86 in complex with wild type HIV-1 protease | | Descriptor: | ACETATE ION, N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{[(4-METHOXYPHENYL)SULFONYL][(2S)-2-METHYLBUTYL]AMINO}PROPYL]-4-OXOHEXANAMIDE, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QI6
 
 | | Crystal structure of protease inhibitor, MIT-2-KB98 in complex with wild type HIV-1 protease | | Descriptor: | N-{(1S,2R)-3-[(1,3-BENZOTHIAZOL-6-YLSULFONYL)(ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYL}-3-HYDROXYBENZAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
