4Q42
 
 | | Crystal structure of Schistosoma mansoni arginase in complex with L-ornithine | | Descriptor: | Arginase, GLYCEROL, L-ornithine, ... | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
3IGG
 
 | | Novel CDK-5 inhibitors - crystal structure of inhibitor EFQ with CDK-2 | | Descriptor: | Cell division protein kinase 2, N-[1-(cis-3-hydroxycyclobutyl)-1H-imidazol-4-yl]-2-(4-methoxyphenyl)acetamide | | Authors: | Pandit, J. | | Deposit date: | 2009-07-27 | | Release date: | 2009-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and cellularly active 4-aminoimidazole inhibitors of cyclin-dependent kinase 5/p25 for the treatment of Alzheimer's disease.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2MSR
 
 | | Solution structure of LEDGF/p75 IBD in complex with MLL1 peptide (140-160) | | Descriptor: | Histone-lysine N-methyltransferase 2A, PC4 and SFRS1-interacting protein | | Authors: | Cermakova, K, Tesina, P, Demeulemeester, J, El Ashkar, S, Mereau, H, Schwaller, J, Rezacova, P, Veverka, V, De Rijck, J. | | Deposit date: | 2014-08-05 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Validation and Structural Characterization of the LEDGF/p75-MLL Interface as a New Target for the Treatment of MLL-Dependent Leukemia.
Cancer Res., 74, 2014
|
|
3KJF
 
 | | Caspase 3 Bound to a covalent inhibitor | | Descriptor: | (3S)-3-({[(5S,10aS)-2-{(2S)-4-carboxy-2-[(phenylacetyl)amino]butyl}-1,3-dioxo-2,3,5,7,8,9,10,10a-octahydro-1H-[1,2,4]triazolo[1,2-a]cinnolin-5-yl]carbonyl}amino)-4-oxopentanoic acid, Caspase-3 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
3KJN
 
 | | Caspase 8 bound to a covalent inhibitor | | Descriptor: | (3S)-3-({[(5S)-2-{2-[(1H-benzimidazol-5-ylcarbonyl)amino]ethyl}-7-(cyclohexylmethyl)-1,3-dioxo-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazin-5-yl]carbonyl}amino)-4-oxopentanoic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Caspase-8 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
3KEC
 
 | | Crystal Structure of Human MMP-13 complexed with a phenyl-2H-tetrazole compound | | Descriptor: | 4-{[({3-[2-(4-methoxybenzyl)-2H-tetrazol-5-yl]phenyl}carbonyl)amino]methyl}benzoic acid, ACETOHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Shieh, H.-S, Pavlovsky, A.G, Collins, B, Schnute, M.E. | | Deposit date: | 2009-10-25 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of (pyridin-4-yl)-2H-tetrazole as a novel scaffold to identify highly selective matrix metalloproteinase-13 inhibitors for the treatment of osteoarthritis.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KEK
 
 | | Crystal Structure of Human MMP-13 complexed with a (pyridin-4-yl)-2H-tetrazole compound | | Descriptor: | CALCIUM ION, Collagenase 3, ZINC ION, ... | | Authors: | Shieh, H.-S, Collins, B, Schnute, M.E. | | Deposit date: | 2009-10-26 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of (pyridin-4-yl)-2H-tetrazole as a novel scaffold to identify highly selective matrix metalloproteinase-13 inhibitors for the treatment of osteoarthritis.
Bioorg.Med.Chem.Lett., 20, 2009
|
|
3L13
 
 | | Crystal Structures of Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors | | Descriptor: | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, [3-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenyl]methanol | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
3KWF
 
 | | human DPP-IV with carmegliptin (S)-1-((2S,3S,11bS)-2-Amino-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-3-yl)-4-fluoromethyl-pyrrolidin-2-one | | Descriptor: | (4S)-1-[(2S,3S,11bS)-2-amino-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-3-yl]-4-(fluoromethyl)pyrrolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Hennig, M, Stihle, M, Thoma, R. | | Deposit date: | 2009-12-01 | | Release date: | 2010-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of carmegliptin: A potent and long-acting dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L3L
 
 | | PARP complexed with A906894 | | Descriptor: | 3-oxo-2-piperidin-4-yl-2,3-dihydro-1H-isoindole-4-carboxamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Park, C.H. | | Deposit date: | 2009-12-17 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and SAR of substituted 3-oxoisoindoline-4-carboxamides as potent inhibitors of poly(ADP-ribose) polymerase (PARP) for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L9M
 
 | | Crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 18 | | Descriptor: | (2S)-N~1~-[5-(3-methyl-1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2PAH
 
 | |
2PB7
 
 | |
2PEV
 
 | | Complex of Aldose Reductase with NADP+ and simaltaneously bound competetive inhibitors Fidarestat and IDD594. Concentration of Fidarestat in soaking solution exceeds concentration of IDD594. | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | Authors: | Petrova, T, Hazemann, I, Cousido, A, Mitschler, A, Ginell, S, Joachimiak, A, Podjarny, A. | | Deposit date: | 2007-04-03 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal packing modifies ligand binding affinity: The case of aldose reductase.
Proteins, 80, 2012
|
|
2PFH
 
 | | Complex of Aldose Reductase with NADP+ and simaltaneously bound competetive inhibitors Fidarestat and IDD594. Concentration of Fidarestat in soaking solution is less than concentration of IDD594. | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | Authors: | Petrova, T, Hazemann, I, Cousido, A, Mitschler, A, Ginell, S, Joachimiak, A, Podjarny, A. | | Deposit date: | 2007-04-05 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Crystal packing modifies ligand binding affinity: The case of aldose reductase.
Proteins, 80, 2012
|
|
2PF8
 
 | | Complex of Aldose Reductase with NADP+ and simaltaneously bound competetive inhibitors Fidarestat and IDD594. Concentration of Fidarestat in soaking solution is equal to concentration of IDD594. | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | Authors: | Petrova, T, Hazemann, I, Cousido, A, Mitschler, A, Ginell, S, Joachimiak, A, Podjarny, A. | | Deposit date: | 2007-04-04 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Crystal packing modifies ligand binding affinity: The case of aldose reductase.
Proteins, 80, 2012
|
|
2PWY
 
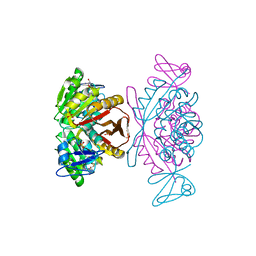 | | Crystal Structure of a m1A58 tRNA methyltransferase | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, tRNA (adenine-N(1)-)-methyltransferase | | Authors: | Barraud, P, Golinelli-Pimpaneau, B, Tisne, C. | | Deposit date: | 2007-05-14 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Thermus thermophilus tRNA m(1)A(58) Methyltransferase and Biophysical Characterization of Its Interaction with tRNA.
J.Mol.Biol., 377, 2008
|
|
3L9N
 
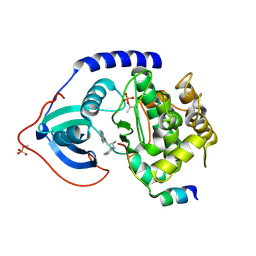 | | crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 27 | | Descriptor: | (2S)-N~1~-[5-(1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L9L
 
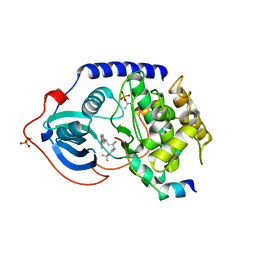 | | Crystal structure of pka with compound 36 | | Descriptor: | 5-[2-({(2S)-2-amino-3-[4-(trifluoromethyl)phenyl]propyl}amino)-1,3-thiazol-5-yl]-1,3-dihydro-2H-indol-2-one, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KEJ
 
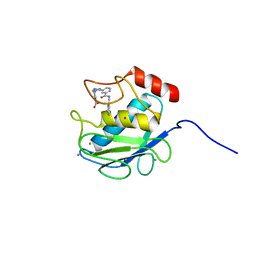 | | Crystal Structure of Human MMP-13 complexed with a (pyridin-4-yl)-2H-tetrazole compound | | Descriptor: | 4-[(5-{2-[(3-fluorobenzyl)carbamoyl]pyridin-4-yl}-2H-tetrazol-2-yl)methyl]benzoic acid, CALCIUM ION, Collagenase 3, ... | | Authors: | Shieh, H.-S, Collins, B, Schnute, M.E. | | Deposit date: | 2009-10-26 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of (pyridin-4-yl)-2H-tetrazole as a novel scaffold to identify highly selective matrix metalloproteinase-13 inhibitors for the treatment of osteoarthritis.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KJQ
 
 | | Caspase 8 with covalent inhibitor | | Descriptor: | (3S)-3-({[(5S,8R)-2-(3-carboxypropyl)-8-(2-{[(4-chlorophenyl)acetyl]amino}ethyl)-1,3-dioxo-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazin-5-yl]carbonyl}amino)-4-oxopentanoic acid, Caspase-8 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
5C6B
 
 | |
5C69
 
 | |
5C14
 
 | | Crystal structure of PECAM-1 D1D2 domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Zhou, D, Paddock, C, Newman, P, Zhu, J. | | Deposit date: | 2015-06-12 | | Release date: | 2016-01-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for PECAM-1 homophilic binding.
Blood, 127, 2016
|
|
5DH5
 
 | | PDE10 complexed with N-[(1-methylpyrazol-4-yl)methyl]-5-[[(1S,2S)-2-(2-pyridyl)cyclopropyl]methoxy]pyrazolo[1,5-a]pyrimidin-7-amine | | Descriptor: | MAGNESIUM ION, N-[(1-methyl-1H-pyrazol-4-yl)methyl]-5-{[(1S,2S)-2-(pyridin-2-yl)cyclopropyl]methoxy}pyrazolo[1,5-a]pyrimidin-7-amine, ZINC ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-08-29 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of pyrazolopyrimidine phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
