6PQL
 
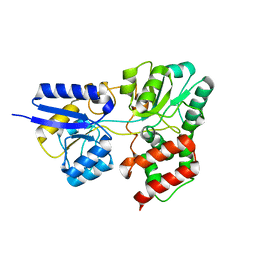 | | SBP RafE in complex with raffinose | | Descriptor: | ABC transporter sugar-binding protein, beta-D-fructofuranose-(2-1)-[alpha-D-galactopyranose-(1-6)]alpha-D-glucopyranose | | Authors: | Meier, E.P.W, Boraston, A.B. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular analysis of an enigmaticStreptococcus pneumoniaevirulence factor: The raffinose-family oligosaccharide utilization system.
J.Biol.Chem., 294, 2019
|
|
7P18
 
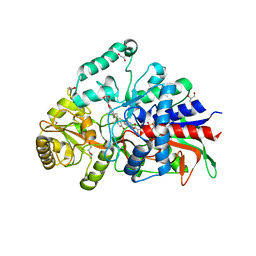 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Sterolibacterium denitrificans in complex with 1,4-androstadiene-3,17-dione | | Descriptor: | 3-oxosteroid 1-dehydrogenase, ANDROSTA-1,4-DIENE-3,17-DIONE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wojcik, P, Mrugala, B, Kurpiewska, K, Szaleniec, M. | | Deposit date: | 2021-07-01 | | Release date: | 2021-07-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure, Mutagenesis, and QM:MM Modeling of 3-Ketosteroid Delta 1 -Dehydrogenase from Sterolibacterium denitrificans ─The Role of a New Putative Membrane-Associated Domain and Proton-Relay System in Catalysis.
Biochemistry, 62, 2023
|
|
8FQC
 
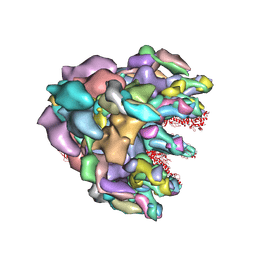 | | Structure of baseplate with receptor binding complex of Agrobacterium phage Milano | | Descriptor: | Baseplate Centerpiece, gp25, Baseplate Central Spike, ... | | Authors: | Sonani, R.R, Leiman, P.G, Wang, F, Kreutzberger, M.A.B, Sebastian, A, Esteves, N.C, Kelly, R.J, Scharf, B, Egelman, E.H. | | Deposit date: | 2023-01-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An extensive disulfide bond network prevents tail contraction in Agrobacterium tumefaciens phage Milano.
Nat Commun, 15, 2024
|
|
4XGO
 
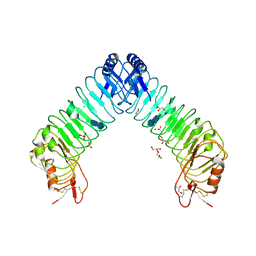 | | Crystal structure of leucine-rich repeat domain of APL1B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Anopheles Plasmodium-responsive Leucine-rich repeat protein 1B, ... | | Authors: | Williams, M, Summers, B, Baxter, R.H.G. | | Deposit date: | 2015-01-01 | | Release date: | 2015-04-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Biophysical Analysis of Anopheles gambiae Leucine-Rich Repeat Proteins APL1A1, APLB and APL1C and Their Interaction with LRIM1.
Plos One, 10, 2015
|
|
5LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
7OQC
 
 | | The U1 part of Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A), Pre-mRNA-processing factor 39, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
5J3X
 
 | | Structure of c-CBL Y371F | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase CBL, ZINC ION | | Authors: | Huang, D.T, Buetow, L, Dou, H. | | Deposit date: | 2016-03-31 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.822 Å) | | Cite: | Casitas B-lineage lymphoma linker helix mutations found in myeloproliferative neoplasms affect conformation.
Bmc Biol., 14, 2016
|
|
7OO6
 
 | | Mechanosensitive channel MscS solubilized with DDM in closed conformation with added lipid | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DODECYL-BETA-D-MALTOSIDE, Mechanosensitive channel of small conductance (MscS) | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5J7C
 
 | |
7P6D
 
 | | Argyrophilic grain disease type 1 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P67
 
 | | Globular glial tauopathy type 2 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P65
 
 | | Progressive supranuclear palsy tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P66
 
 | | Globular glial tauopathy type 1 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P6B
 
 | | Limbic-predominant neuronal inclusion body 4R tauopathy type 1b tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P6C
 
 | | Limbic-predominant neuronal inclusion body 4R tauopathy type 2 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
6VCB
 
 | | Cryo-EM structure of the Glucagon-like peptide-1 receptor in complex with G protein, GLP-1 peptide and a positive allosteric modulator | | Descriptor: | 1-[(1R)-1-(2,6-dichloro-3-methoxyphenyl)ethyl]-6-{2-[(2R)-piperidin-2-yl]phenyl}-1H-benzimidazole, Glucagon-like peptide 1, Glucagon-like peptide 1 receptor, ... | | Authors: | Sun, B, Feng, D, Bueno, A, Kobilka, B, Sloop, K. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into probe-dependent positive allosterism of the GLP-1 receptor.
Nat.Chem.Biol., 16, 2020
|
|
6MS3
 
 | | Crystal structure of the GH43 protein BlXynB mutant (K247S) from Bacillus licheniformis | | Descriptor: | CALCIUM ION, GLYCEROL, Glycoside Hydrolase Family 43, ... | | Authors: | Zanphorlin, L.M, Morais, M.A.B, Diogo, J.A, Murakami, M.T. | | Deposit date: | 2018-10-16 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design combined with evolutionary diversity led to the discovery of the xylose-releasing exo-xylanase activity in the glycoside hydrolase family 43.
Biotechnol. Bioeng., 116, 2019
|
|
4XOC
 
 | | Crystal structure of the FimH lectin domain from E.coli F18 in complex with heptyl alpha-D-mannopyrannoside | | Descriptor: | FimH protein, heptyl alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Sauer, M.M, Navarra, G, Ernst, B, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
8E8V
 
 | |
1BW9
 
 | | PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NAD+ AND PHENYLPYRUVATE | | Descriptor: | 1,2-ETHANEDIOL, 3-PHENYLPYRUVIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Vanhooke, J.L, Thoden, J.B, Brunhuber, N.M.W, Blanchard, J.L, Holden, H.M. | | Deposit date: | 1998-10-01 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phenylalanine dehydrogenase from Rhodococcus sp. M4: high-resolution X-ray analyses of inhibitory ternary complexes reveal key features in the oxidative deamination mechanism.
Biochemistry, 38, 1999
|
|
7O2I
 
 | | METTL3-METTL14 heterodimer bound to the SAM competitive small molecule inhibitor STM2457 | | Descriptor: | DIMETHYL SULFOXIDE, N6-adenosine-methyltransferase catalytic subunit, N6-adenosine-methyltransferase non-catalytic subunit, ... | | Authors: | Pilka, E.S, Blackaby, W, Hardick, D, Harper, C, Hewstone, D, Ridgill, M, Rotty, B, Rausch, O. | | Deposit date: | 2021-03-30 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia.
Nature, 593, 2021
|
|
6P9A
 
 | | HIV-1 Protease multiple mutant PRS5B with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 Protease, PHOSPHATE ION | | Authors: | Kneller, D.W, Agniswamy, J, Weber, I.T. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Highly drug-resistant HIV-1 protease reveals decreased intra-subunit interactions due to clusters of mutations.
Febs J., 287, 2020
|
|
8CUU
 
 | |
6XVC
 
 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 1 | | Descriptor: | (4~{R})-4-[(1~{R})-1-[7-(3-methyl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)quinolin-5-yl]oxyethyl]pyrrolidin-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.098 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6TCV
 
 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 in complex with Man9GlcNAc2Asn substrate | | Descriptor: | ASPARAGINE, Endo-beta-N-acetylglucosaminidase F1, GLYCEROL, ... | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-11-06 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.311 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
