8S98
 
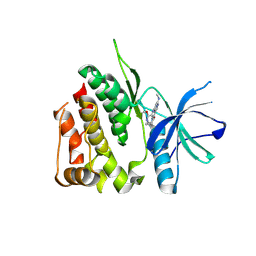 | | Crystal structure of the TYK2 pseudokinase domain in complex with compound 8 | | Descriptor: | (8S)-N-cyclopropyl-5-[(2-methoxypyridin-3-yl)amino]-7-(methylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Carriero, S, Mondal, S, Abel, R, Ashwell, M, Blanchette, H, Boyles, N, Cartwright, M, Collis, A, Feng, S, Ghanakota, P, Harriman, G.C, Hosagrahara, V, Kaila, N, Kapeller, R, Rafi, S, Romero, D.L, Tarantino, P, Timaniya, J, Wester, R.T, Westlin, W, Srivastava, B, Miao, W, Tummino, P, McElwee, J.J, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2023-03-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of a Potent and Selective Tyrosine Kinase 2 Inhibitor: TAK-279.
J.Med.Chem., 66, 2023
|
|
8S9B
 
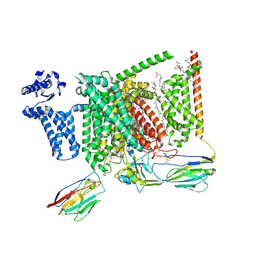 | | Cryo-EM structure of Nav1.7 with LCM | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8S9C
 
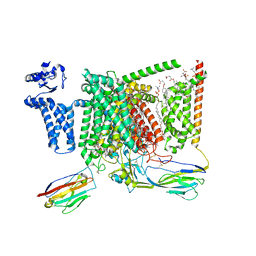 | | Cryo-EM structure of Nav1.7 with CBZ | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8SBD
 
 | | Cryo-EM structure of insulin amyloid-like fibril that is composed of two antiparallel protofilaments | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Wang, L.W, Hall, C, Uchikawa, E, Chen, D.L, Choi, E, Zhang, X.W, Bai, X.C. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of insulin fibrillation.
Sci Adv, 9, 2023
|
|
6P4Y
 
 | |
5T9R
 
 | | Structure of rabbit RyR1 (Ca2+-only dataset, class 3) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TB0
 
 | | Structure of rabbit RyR1 (EGTA-only dataset, all particles) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
6P67
 
 | | Crystal Structure of a Complex of human IL-7Ralpha with an anti-IL-7Ralpha 2B8 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-IL-7R 2B8 Fab heavy chain, ... | | Authors: | Walsh, S.T.R, Kashi, L, Kohnhorst, C.L. | | Deposit date: | 2019-06-03 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | New anti-IL-7R alpha monoclonal antibodies show efficacy against T cell acute lymphoblastic leukemia in pre-clinical models.
Leukemia, 34, 2020
|
|
6P50
 
 | | Crystal Structure of a Complex of human IL-7Ralpha with an anti-IL-7Ralpha Fab 4A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-7 receptor subunit alpha, anti-IL-7R 4A10 Fab heavy chain, ... | | Authors: | Walsh, S.T.R, Kashi, L, Kohnhorst, C.L. | | Deposit date: | 2019-05-29 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | New anti-IL-7R alpha monoclonal antibodies show efficacy against T cell acute lymphoblastic leukemia in pre-clinical models.
Leukemia, 34, 2020
|
|
5TAS
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/EGTA dataset, class 1) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
6P2J
 
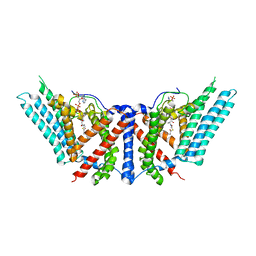 | | Dimeric structure of ACAT1 | | Descriptor: | S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name), Sterol O-acyltransferase 1 | | Authors: | Yan, N, Qian, H.W. | | Deposit date: | 2019-05-21 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for catalysis and substrate specificity of human ACAT1.
Nature, 581, 2020
|
|
5M7G
 
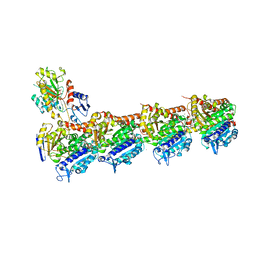 | | Tubulin-MTD147 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(2,6-dimorpholin-4-ylpyridin-4-yl)-4-(trifluoromethyl)pyridin-2-amine, CALCIUM ION, ... | | Authors: | Bohnacker, T, Prota, A.E, Steinmetz, M.O, Wymann, M.P. | | Deposit date: | 2016-10-27 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
6P4V
 
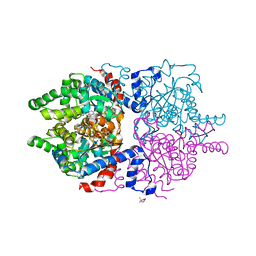 | |
5TA3
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 2) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TB2
 
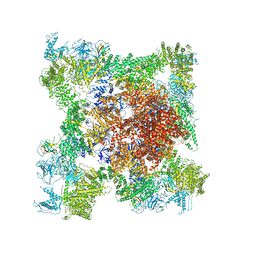 | | Structure of rabbit RyR1 (EGTA-only dataset, class 2) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TDE
 
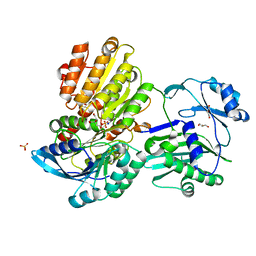 | | TEV Cleaved Human ATP Citrate Lyase Bound to Citrate | | Descriptor: | ATP-citrate synthase, CITRATE ANION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Hu, J, Fraser, M.E. | | Deposit date: | 2016-09-19 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of hydroxycitrate to human ATP-citrate lyase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5MAR
 
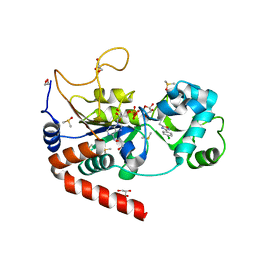 | | Structure of human SIRT2 in complex with 1,2,4-Oxadiazole inhibitor and ADP ribose. | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(4-chlorophenyl)-1,2,4-oxadiazol-5-yl]propan-1-ol, ACETATE ION, ... | | Authors: | Moniot, S, Steegborn, C. | | Deposit date: | 2016-11-04 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Development of 1,2,4-Oxadiazoles as Potent and Selective Inhibitors of the Human Deacetylase Sirtuin 2: Structure-Activity Relationship, X-ray Crystal Structure, and Anticancer Activity.
J. Med. Chem., 60, 2017
|
|
5M7E
 
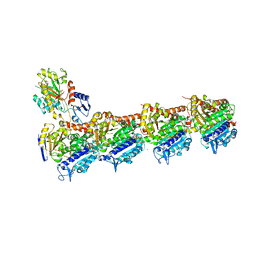 | | Tubulin-BKM120 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[2,6-di(morpholin-4-yl)pyrimidin-4-yl]-4-(trifluoromethyl)pyridin-2-amine, CALCIUM ION, ... | | Authors: | Bohnacker, T, Prota, A.E, Steinmetz, M.O, Wymann, M.P. | | Deposit date: | 2016-10-27 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
5TAN
 
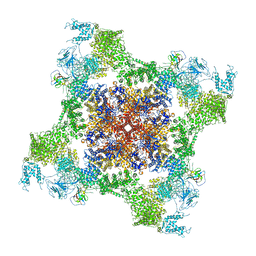 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 3) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
6P96
 
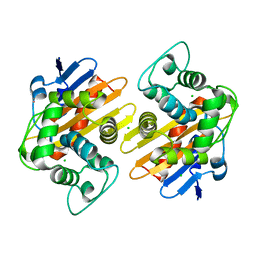 | | OXA-48 carbapanemase, apo form | | Descriptor: | Beta-lactamase, CADMIUM ION, CALCIUM ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Mechanism of Carbapenemase Activity of the OXA-48 beta-Lactamase.
Antimicrob.Agents Chemother., 63, 2019
|
|
6P9C
 
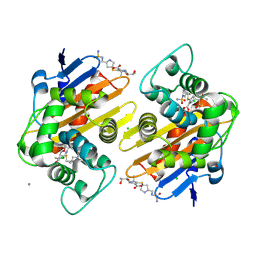 | | OXA-48 carbapanemase, doripenem complex | | Descriptor: | (4R,5S)-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-3-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CADMIUM ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-07 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Mechanism of Carbapenemase Activity of the OXA-48 beta-Lactamase.
Antimicrob.Agents Chemother., 63, 2019
|
|
6P79
 
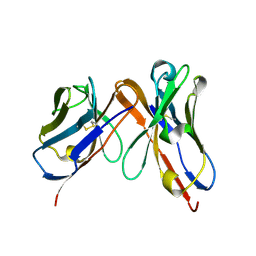 | | Engineered single chain antibody C9+C14 ScFv | | Descriptor: | Engineered antibody heavy chain, Engineered antibody light chain | | Authors: | Zhang, Y, Li, W, Marshall, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Computer-based Engineering of Thermostabilized Antibody Fragments.
Aiche J, 66, 2020
|
|
6P3T
 
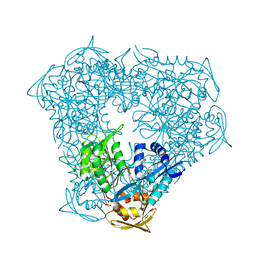 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT449 | | Descriptor: | AMMONIUM ION, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2019-05-24 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the Robustness of Inhibitors of Tuberculosis Aminoglycoside Resistance Enzyme Eis by Mutagenesis.
Acs Infect Dis., 5, 2019
|
|
5M7J
 
 | | Blastochloris viridis photosynthetic reaction center structure using best crystal approach | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Sharma, A.S, Johansson, L, Dunevall, E, Wahlgren, W.Y, Neutze, R, Katona, G. | | Deposit date: | 2016-10-28 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Asymmetry in serial femtosecond crystallography data.
Acta Crystallogr A Found Adv, 73, 2017
|
|
8RCW
 
 | | Crystal structure of the Mycobacterium tuberculosis regulator VirS (N-terminal fragment 4-208) in complex with the lead compound SMARt751 | | Descriptor: | 4,4,4-tris(fluoranyl)-1-[4-(4-fluorophenyl)piperidin-1-yl]butan-1-one, HTH-type transcriptional regulator VirS | | Authors: | Grosse, C, Sigoillot, M, Megalizzi, V, Tanina, A, Willand, N, Baulard, A.R, Wintjens, R. | | Deposit date: | 2023-12-07 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis VirS regulator reveals its interaction with the lead compound SMARt751.
J.Struct.Biol., 216, 2024
|
|
